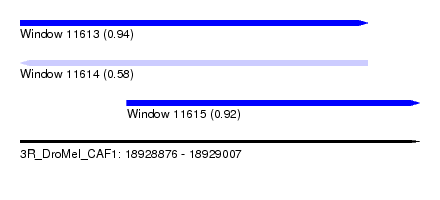
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,928,876 – 18,929,007 |
| Length | 131 |
| Max. P | 0.943297 |

| Location | 18,928,876 – 18,928,990 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 84.64 |
| Mean single sequence MFE | -33.78 |
| Consensus MFE | -21.72 |
| Energy contribution | -22.16 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.90 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.943297 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18928876 114 + 27905053 UAUGCCUAUUUUUUCGACCUGAGCUGUAUGGCUAUAUCCUCAAGCGCAAAUAUUAUAUGUACAACAUAUAUUUCGCUGGGGACAGUAAAGUGUUCUAAUGCCACACACAGGCAA ..(((((...........(((((((....))))...((((((.(((.(((((...((((.....)))))))))))))))))))))....((((.........))))..))))). ( -30.10) >DroSec_CAF1 39989 114 + 1 AAUGCCUAUUUUUUCGACCUCAGCUGUAUGGCUACAUCCUCAAGCGCGAGUUUUAUAUGUACAACAUAUAUUUCGCUGGGGACAGCAAAGUGUUCUAAUGCGACACAAAGGCAA ..(((((...............((((((((....)))(((((...(((((...((((((.....)))))).)))))))))))))))...(((((.......)))))..))))). ( -34.60) >DroSim_CAF1 39966 114 + 1 AAUGCCUAUUUUUUUGACCUGCGCUGUAUGGCUCUAUCCUCAAGCGCGAGUUUUAUAUGUACAACAUAUAUUUCGCUGGGGACAGCAAAGUGUUCUAAUGCGACACAAAGGCAA ..(((((.............(((((....((......))...)))))(((...((((((.....)))))).)))((((....))))...(((((.......)))))..))))). ( -35.20) >DroEre_CAF1 40132 99 + 1 UGUGCCUAUUUUUGCGACUUGAGCUGUGUGG---------------CAAGUGUUAUAUGCACAACAUAUACUUAGCUGGGGACAGCAAAGUGUUCUAAUGCGACACAAAGGAAA ....(((......((.......))(((((.(---------------((.((((.....)))).....((((((.((((....)))).)))))).....))).))))).)))... ( -32.60) >DroYak_CAF1 41237 113 + 1 UGUGCCUAUUUUUUUGGCCUCAGCUGUGUGGCCAUAUUCUCG-GCGCAAGUAUUAUAUGUACAACAUAUAUUUAGCUGGGGACAGCAAAGUGUUCUAGUGCGACACAAAGGCAA ..(((((.......(((((.((....)).)))))........-((((..((((.....)))).....((((((.((((....)))).))))))....)))).......))))). ( -36.40) >consensus UAUGCCUAUUUUUUCGACCUGAGCUGUAUGGCUAUAUCCUCAAGCGCAAGUAUUAUAUGUACAACAUAUAUUUCGCUGGGGACAGCAAAGUGUUCUAAUGCGACACAAAGGCAA ..(((((...............(((....))).....................((((((.....))))))....((((....))))...(((((.......)))))..))))). (-21.72 = -22.16 + 0.44)


| Location | 18,928,876 – 18,928,990 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 84.64 |
| Mean single sequence MFE | -28.84 |
| Consensus MFE | -18.88 |
| Energy contribution | -19.48 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.584927 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18928876 114 - 27905053 UUGCCUGUGUGUGGCAUUAGAACACUUUACUGUCCCCAGCGAAAUAUAUGUUGUACAUAUAAUAUUUGCGCUUGAGGAUAUAGCCAUACAGCUCAGGUCGAAAAAAUAGGCAUA .((((((((((((((..((((....)))).((((((.((((((((((((((.....)))).)))))).)))).).)))))..)))))))...((.....))....))))))).. ( -32.90) >DroSec_CAF1 39989 114 - 1 UUGCCUUUGUGUCGCAUUAGAACACUUUGCUGUCCCCAGCGAAAUAUAUGUUGUACAUAUAAAACUCGCGCUUGAGGAUGUAGCCAUACAGCUGAGGUCGAAAAAAUAGGCAUU .(((((..(((((......).))))(((((((....))))))).((((((.....))))))....(((.((((.((..((((....)))).)).)))))))......))))).. ( -29.60) >DroSim_CAF1 39966 114 - 1 UUGCCUUUGUGUCGCAUUAGAACACUUUGCUGUCCCCAGCGAAAUAUAUGUUGUACAUAUAAAACUCGCGCUUGAGGAUAGAGCCAUACAGCGCAGGUCAAAAAAAUAGGCAUU .(((((..(((((......).))))(((((((....))))))).((((((.....))))))..((..(((((((.((......)).)).)))))..)).........))))).. ( -30.20) >DroEre_CAF1 40132 99 - 1 UUUCCUUUGUGUCGCAUUAGAACACUUUGCUGUCCCCAGCUAAGUAUAUGUUGUGCAUAUAACACUUG---------------CCACACAGCUCAAGUCGCAAAAAUAGGCACA .......((((((((((......((((.((((....)))).)))).......)))).........(((---------------(.((.........)).)))).....)))))) ( -22.42) >DroYak_CAF1 41237 113 - 1 UUGCCUUUGUGUCGCACUAGAACACUUUGCUGUCCCCAGCUAAAUAUAUGUUGUACAUAUAAUACUUGCGC-CGAGAAUAUGGCCACACAGCUGAGGCCAAAAAAAUAGGCACA .((((((((.(.((((............((((....))))....((((((.....)))))).....)))))-))).....(((((.((....)).))))).......))))).. ( -29.10) >consensus UUGCCUUUGUGUCGCAUUAGAACACUUUGCUGUCCCCAGCGAAAUAUAUGUUGUACAUAUAAAACUUGCGCUUGAGGAUAUAGCCAUACAGCUCAGGUCGAAAAAAUAGGCAUA .(((((.(((((.((..........(((((((....))))))).((((((.....)))))).....................)).)))))((....)).........))))).. (-18.88 = -19.48 + 0.60)


| Location | 18,928,911 – 18,929,007 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 79.75 |
| Mean single sequence MFE | -25.38 |
| Consensus MFE | -17.28 |
| Energy contribution | -17.78 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.918272 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18928911 96 + 27905053 AUCCUCAAGCGCAAAUAUUAUAUGUACAACAUAUAUUUCGCUGGGGACAGUAAAGUGUUCU-AAUGCCACACACAGGCAAUUGCCGACGCCUUGUUU .((((((.(((.(((((...((((.....))))))))))))))))))...(((.(((((.(-((((((.......))).))))..))))).)))... ( -23.30) >DroSec_CAF1 40024 96 + 1 AUCCUCAAGCGCGAGUUUUAUAUGUACAACAUAUAUUUCGCUGGGGACAGCAAAGUGUUCU-AAUGCGACACAAAGGCAAUUGCCGUCGCCUUGUUU .....((((.(((((...((((((.....))))))...)((((....))))...(((((..-.....)))))...(((....))).))))))))... ( -30.50) >DroSim_CAF1 40001 96 + 1 AUCCUCAAGCGCGAGUUUUAUAUGUACAACAUAUAUUUCGCUGGGGACAGCAAAGUGUUCU-AAUGCGACACAAAGGCAAUUGCCGCCGCCUUGUUU .....((((.(((.((..((((((.....))))))....((((....))))...(((((..-.....)))))...(((....))))))))))))... ( -29.20) >DroEre_CAF1 40163 85 + 1 -----------CAAGUGUUAUAUGCACAACAUAUACUUAGCUGGGGACAGCAAAGUGUUCU-AAUGCGACACAAAGGAAAUUGCCGCCGCCUUGUUU -----------...(((((....(((......((((((.((((....)))).))))))...-..)))))))).((((............)))).... ( -22.60) >DroYak_CAF1 41272 95 + 1 AUUCUCG-GCGCAAGUAUUAUAUGUACAACAUAUAUUUAGCUGGGGACAGCAAAGUGUUCU-AGUGCGACACAAAGGCAAUUGCCGCCGCCUUGUUU ......(-((((..........(((((.....((((((.((((....)))).))))))...-.))))).......(((....)))).))))...... ( -28.80) >DroAna_CAF1 41967 80 + 1 --------------UUUCUAAGUGUAUUUUAUAUAUUUAACUGGAGACAACAAAGUUUCCUUUAUGUGACACACAGGCAAUUGCCAGCGCCUUG--- --------------.......((((...(((((((.......((((((......))))))..))))))).)))).(((....))).........--- ( -17.90) >consensus AUCCUCA_GCGCAAGUUUUAUAUGUACAACAUAUAUUUAGCUGGGGACAGCAAAGUGUUCU_AAUGCGACACAAAGGCAAUUGCCGCCGCCUUGUUU ..................((((((.....))))))....((((....))))...(((((........)))))...(((....)))............ (-17.28 = -17.78 + 0.50)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:39:24 2006