
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,914,286 – 18,914,377 |
| Length | 91 |
| Max. P | 0.986270 |

| Location | 18,914,286 – 18,914,377 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 76.58 |
| Mean single sequence MFE | -25.29 |
| Consensus MFE | -9.49 |
| Energy contribution | -9.82 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.22 |
| Structure conservation index | 0.38 |
| SVM decision value | 2.04 |
| SVM RNA-class probability | 0.986270 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18914286 91 + 27905053 CAAUAACGCCGUUAGCCGUGCUCCAUCUGCAUUUAUCAUAACUUUUGUGUGUUCUUUGCUUUGAUGGCG---CUGCU---GUGGCUC--G-CGGUUAU-UG (((((..(((((.(((((.((.(((((.(((.....((((.(....))))).....)))...))))).)---)....---.))))).--)-)))))))-)) ( -30.80) >DroPse_CAF1 28495 90 + 1 CAAUAAUGCCUUUAGCUAUGCUCCAUCUGCAUUUAUCAUAACUUUUGUGUUUUAUUUGCUUUGAUGGCG---UUGCG---UAGCCCC--C-AGGCUGU-U- .......((((...(((((((.(((((.(((..((.((((.....))))...))..)))...)))))..---..)))---))))...--.-))))...-.- ( -28.00) >DroGri_CAF1 28766 95 + 1 CAAUAAUGCUGCUAGCUAUGCUCCAUCUGCAUUUAUCAUAACUUUCCUCUUUUAUUUGCCUUGAUGGCAGCAUUGCC---ACCAGUC--U-UGUUUUUCUC .(((((.((((...((.((((((((((.(((..((.................))..)))...))))).))))).)).---..)))).--)-))))...... ( -23.83) >DroMoj_CAF1 29863 94 + 1 CAAUAACGCUGCCAGCUAUACUCCAUCUGCAUUUAUCAUAACUUUAGUCCUUUAUUUGCCUUGAUGGCAACACUGCC---ACAGGUC--G-CAUUUGU-UG ((((((.((.(((...............(((..((.....((....))....))..)))..((.(((((....))))---)))))).--)-)..))))-)) ( -17.20) >DroAna_CAF1 26205 96 + 1 CAAUAAUGCCGUUAGCCGUGCUCCAUCUGCAUUUAUCAUAACUUUUGUGUGUUAUUUGCUUUGAUGGCA---UCGCCCUCUUCGCCCUCG-GGGCCAU-UG .....(((((((((((.((((.......)))).....(((((........)))))..)))..)))))))---).(((((..........)-))))...-.. ( -23.30) >DroPer_CAF1 28565 91 + 1 CAAUAAUGCCUUUAGCUAUGCUCCAUCUGCAUUUAUCAUAACUUUUGUGUUUUAUUUGCUUUGAUGGCG---UUGCG---UAGCCCC--CAAGGCUGU-U- .......(((((..(((((((.(((((.(((..((.((((.....))))...))..)))...)))))..---..)))---))))...--.)))))...-.- ( -28.60) >consensus CAAUAAUGCCGUUAGCUAUGCUCCAUCUGCAUUUAUCAUAACUUUUGUGUUUUAUUUGCUUUGAUGGCA___UUGCC___UAGGCCC__G_AGGCUGU_UG ..............((......(((((.(((..((.....((....))....))..)))...))))).......))......................... ( -9.49 = -9.82 + 0.33)


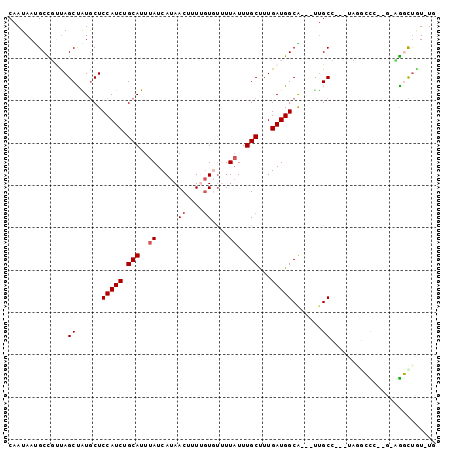
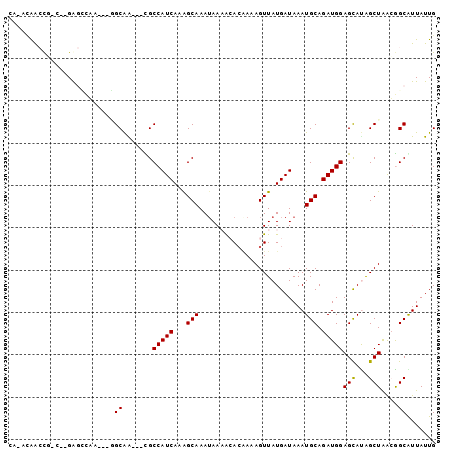
| Location | 18,914,286 – 18,914,377 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 76.58 |
| Mean single sequence MFE | -22.70 |
| Consensus MFE | -8.88 |
| Energy contribution | -8.74 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.39 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.544128 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18914286 91 - 27905053 CA-AUAACCG-C--GAGCCAC---AGCAG---CGCCAUCAAAGCAAAGAACACACAAAAGUUAUGAUAAAUGCAGAUGGAGCACGGCUAACGGCGUUAUUG ((-(((((((-.--.((((..---....(---(.(((((...(((...(((........)))........))).))))).))..))))..)))..)))))) ( -21.20) >DroPse_CAF1 28495 90 - 1 -A-ACAGCCU-G--GGGGCUA---CGCAA---CGCCAUCAAAGCAAAUAAAACACAAAAGUUAUGAUAAAUGCAGAUGGAGCAUAGCUAAAGGCAUUAUUG -.-...((((-.--..(((((---.((..---..(((((...(((.((..(((......)))...))...))).))))).)).)))))..))))....... ( -23.50) >DroGri_CAF1 28766 95 - 1 GAGAAAAACA-A--GACUGGU---GGCAAUGCUGCCAUCAAGGCAAAUAAAAGAGGAAAGUUAUGAUAAAUGCAGAUGGAGCAUAGCUAGCAGCAUUAUUG ..........-.--(.(((.(---(((.(((((.(((((...(((.........................))).)))))))))).)))).))))....... ( -25.01) >DroMoj_CAF1 29863 94 - 1 CA-ACAAAUG-C--GACCUGU---GGCAGUGUUGCCAUCAAGGCAAAUAAAGGACUAAAGUUAUGAUAAAUGCAGAUGGAGUAUAGCUGGCAGCGUUAUUG ..-...((((-(--..((...---(((.(((...(((((...(((.......(((....)))........))).)))))..))).)))))..))))).... ( -21.16) >DroAna_CAF1 26205 96 - 1 CA-AUGGCCC-CGAGGGCGAAGAGGGCGA---UGCCAUCAAAGCAAAUAACACACAAAAGUUAUGAUAAAUGCAGAUGGAGCACGGCUAACGGCAUUAUUG ..-((.((((-(.........).)))).)---).(((((...(((.(((((........)))))......))).))))).((.((.....))))....... ( -21.30) >DroPer_CAF1 28565 91 - 1 -A-ACAGCCUUG--GGGGCUA---CGCAA---CGCCAUCAAAGCAAAUAAAACACAAAAGUUAUGAUAAAUGCAGAUGGAGCAUAGCUAAAGGCAUUAUUG -.-...(((((.--..(((((---.((..---..(((((...(((.((..(((......)))...))...))).))))).)).))))).)))))....... ( -24.00) >consensus CA_ACAACCG_C__GAGCCAA___GGCAA___CGCCAUCAAAGCAAAUAAAACACAAAAGUUAUGAUAAAUGCAGAUGGAGCAUAGCUAACGGCAUUAUUG .........................((.......(((((...(((.........................))).)))))(((...)))....))....... ( -8.88 = -8.74 + -0.14)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:39:15 2006