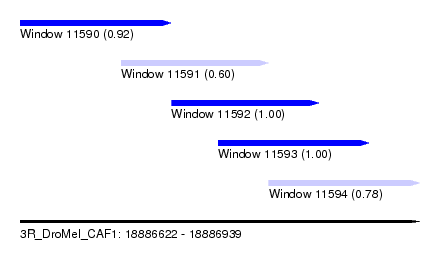
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,886,622 – 18,886,939 |
| Length | 317 |
| Max. P | 0.999992 |

| Location | 18,886,622 – 18,886,742 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.42 |
| Mean single sequence MFE | -53.12 |
| Consensus MFE | -49.66 |
| Energy contribution | -50.58 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.917636 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18886622 120 + 27905053 CAUGGUAGUAGUCCGGCUGCGAGUCCGUGCAGGAGUAGGUGUUCCGACUGGUGCUCGAGCUGCGCGGAUGGAACUCGCAGCUGGGCGAGGUGCUCGCACAGCGCUGCAGCGAUUUCUUCU ....(((((.((((((((((((((((((((((((((((((......)))..)))))...))))))))).....)))))))))))))(..(((....)))..))))))............. ( -55.80) >DroSec_CAF1 886 120 + 1 CAUGGUAGUAGUCCGGCUGCGAGUCCGUGCAGGAGUAGGUGUUCCGACUGGUGCUCGAGCUGCGCGGAUGGAACUCGCAGCUGGGCGAGGUGCUCGCACAGCGCUGCAGCGACUUCUUCU ..........((((((((((((((((((((((((((((((......)))..)))))...))))))))).....)))))))))))))((((((((.(((......)))))).))))).... ( -57.50) >DroSim_CAF1 531 119 + 1 CAUG-UAGUAGUCCGGCCGCGAGUCCGUGCAGGAGUAGGUGUUCCGACUCGUGCUCGAGCUGCACCAUUGGAACUUGCAGCUGGGGGAGGUGCUAGCACAGCGGCGCAGCGACUUCUUCU ....-......((((((.(((((((((((((((((((.(.((....)).).)))))...))))))....)).)))))).))))))(((((((((.((......))..))).))))))... ( -46.80) >DroEre_CAF1 665 120 + 1 CAUGGUAGUAGUCCGGCUGCGAGUCCGUGCAGGAGUAGGUGUUCCGACUGGUGCUCGAGCUGCGCGGAUGGAACUCACAGCUGGGCGAGGUGCUGGCACAGCGCUGCAGCGACUUCUUCU ..........(((((((((.((((((((((((((((((((......)))..)))))...))))))))).....))).)))))))))(((((((((((.....))..)))).))))).... ( -52.10) >DroYak_CAF1 528 120 + 1 CAUGAUAGUAGUCCGGCUGGGAGUCCGUGCAGGAGUAGGUGUUCCGACUGGUGCUCGAGCUGCGCGGAUGGAACUCGCAGCUGGGCGAGGUGCUGGCACAGCGCUGCAGCGACUUCUUCU ..........(((((((((.((((((((((((((((((((......)))..)))))...))))))))).....))).)))))))))(((((((((((.....))..)))).))))).... ( -53.40) >consensus CAUGGUAGUAGUCCGGCUGCGAGUCCGUGCAGGAGUAGGUGUUCCGACUGGUGCUCGAGCUGCGCGGAUGGAACUCGCAGCUGGGCGAGGUGCUCGCACAGCGCUGCAGCGACUUCUUCU ..........((((((((((((((((((((((((((((((......)))..)))))...)))))))).....))))))))))))))((((((((.(((......)))))).))))).... (-49.66 = -50.58 + 0.92)

| Location | 18,886,702 – 18,886,819 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.08 |
| Mean single sequence MFE | -43.50 |
| Consensus MFE | -37.96 |
| Energy contribution | -37.56 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.595362 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18886702 117 + 27905053 CUGGGCGAGGUGCUCGCACAGCGCUGCAGCGAUUUCUUCUUGGCCCGCUGAUCGCU---CCACACAUUGAUCACCGAAAUAUCCGGUGGGGUGUCCUUCUUCAUGUACUUCCCCACCACG .((((.(((((((......((((...(((((...((.....))..)))))..))))---..((((.....((((((.......)))))).))))..........)))))))))))..... ( -38.20) >DroSec_CAF1 966 120 + 1 CUGGGCGAGGUGCUCGCACAGCGCUGCAGCGACUUCUUCUUGGCCCGCUGGUCGCUCGUCCACACAUUGAUCGCCGAAAUCUCCGGUGGGGUGUCCUUCUUCAUGUACUUCCCCACCACG .(((((((((((((.....)))))..(((((.((.......))..)))))....))))))))......................(((((((.((.(........).))..)))))))... ( -41.00) >DroSim_CAF1 610 120 + 1 CUGGGGGAGGUGCUAGCACAGCGGCGCAGCGACUUCUUCUUGGCCCGUUGGUCGCUAGUCCACACAUUGAUCGCCGAAAUCUCCGGUGGGGUGUCCUUCUUCAUGUACUUCCCCACCACG .(((((((((..(((((.((((((.((...((.....))...))))))))...)))))..)((((.....((((((.......)))))).)))).............))))))))..... ( -45.60) >DroEre_CAF1 745 120 + 1 CUGGGCGAGGUGCUGGCACAGCGCUGCAGCGACUUCUUCUUGGCACGCUGGUCGCUCGUCCACACAUUGAUCACCGGAAUCUCCGGUGGGGUGUCCUUCUUCAUGUACUUCCCCACCACG .((((.(((((((.(((.(((((.(((...((.....))...)))))))))))........((((.....(((((((.....))))))).))))..........)))))))))))..... ( -46.10) >DroYak_CAF1 608 120 + 1 CUGGGCGAGGUGCUGGCACAGCGCUGCAGCGACUUCUUCUUGGCACGCUGAUCGCUCGUCCACACAUUGAUCACCGGAAUCUCCGGUGGGGUGUCCUUCUUCAUGUACUUCCCCACCACG .((((.(((((((.((.((((((...(((((.((.......))..)))))..)))).))))((((.....(((((((.....))))))).))))..........)))))))))))..... ( -46.60) >consensus CUGGGCGAGGUGCUCGCACAGCGCUGCAGCGACUUCUUCUUGGCCCGCUGGUCGCUCGUCCACACAUUGAUCACCGAAAUCUCCGGUGGGGUGUCCUUCUUCAUGUACUUCCCCACCACG .((((.(((((((......((((...(((((...((.....))..)))))..)))).....((((.....((((((.......)))))).))))..........)))))))))))..... (-37.96 = -37.56 + -0.40)


| Location | 18,886,742 – 18,886,859 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.08 |
| Mean single sequence MFE | -47.26 |
| Consensus MFE | -47.38 |
| Energy contribution | -46.90 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.52 |
| Structure conservation index | 1.00 |
| SVM decision value | 5.68 |
| SVM RNA-class probability | 0.999992 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18886742 117 + 27905053 UGGCCCGCUGAUCGCU---CCACACAUUGAUCACCGAAAUAUCCGGUGGGGUGUCCUUCUUCAUGUACUUCCCCACCACGCCCUCCUUGUGCUGGGACUUCUCCUUGUUCCGGCGCGAGA .(((.((.((((((..---........)))))).))........(((((((.((.(........).))..)))))))..)))...(((((((((((((........))))))))))))). ( -47.30) >DroSec_CAF1 1006 120 + 1 UGGCCCGCUGGUCGCUCGUCCACACAUUGAUCGCCGAAAUCUCCGGUGGGGUGUCCUUCUUCAUGUACUUCCCCACCACGCCCUCCUUGUGCUGGGACUUCUCCUUGUUCCGGCGCGAGA .(((....(((.((.(((.........))).)))))........(((((((.((.(........).))..)))))))..)))...(((((((((((((........))))))))))))). ( -46.00) >DroSim_CAF1 650 120 + 1 UGGCCCGUUGGUCGCUAGUCCACACAUUGAUCGCCGAAAUCUCCGGUGGGGUGUCCUUCUUCAUGUACUUCCCCACCACGCCCUCCUUGUGCUGGGACUUCUCCUUGUUCCGGCGCGAGA .(((.((.((((((...((....))..)))))).))........(((((((.((.(........).))..)))))))..)))...(((((((((((((........))))))))))))). ( -46.00) >DroEre_CAF1 785 120 + 1 UGGCACGCUGGUCGCUCGUCCACACAUUGAUCACCGGAAUCUCCGGUGGGGUGUCCUUCUUCAUGUACUUCCCCACCACGCCCUCCUUGUGCUGGGACUUCUCCUUGUUCCGGCGCGAGA .(((...(((((.(.(((.........))).)))))).......(((((((.((.(........).))..)))))))..)))...(((((((((((((........))))))))))))). ( -48.40) >DroYak_CAF1 648 120 + 1 UGGCACGCUGAUCGCUCGUCCACACAUUGAUCACCGGAAUCUCCGGUGGGGUGUCCUUCUUCAUGUACUUCCCCACCACGCCCUCCUUGUGCUGGGACUUCUCCUUGUUCCGGCGCGAGA .(((.((.((((((...((....))..)))))).))........(((((((.((.(........).))..)))))))..)))...(((((((((((((........))))))))))))). ( -48.60) >consensus UGGCCCGCUGGUCGCUCGUCCACACAUUGAUCACCGAAAUCUCCGGUGGGGUGUCCUUCUUCAUGUACUUCCCCACCACGCCCUCCUUGUGCUGGGACUUCUCCUUGUUCCGGCGCGAGA .(((.((.((((((.............)))))).))........(((((((.((.(........).))..)))))))..)))...(((((((((((((........))))))))))))). (-47.38 = -46.90 + -0.48)


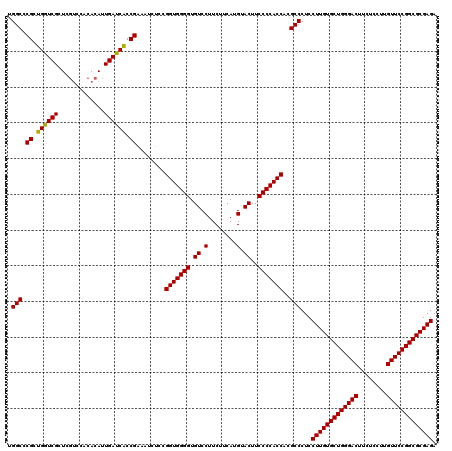
| Location | 18,886,779 – 18,886,899 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.67 |
| Mean single sequence MFE | -47.40 |
| Consensus MFE | -47.34 |
| Energy contribution | -47.34 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.80 |
| Structure conservation index | 1.00 |
| SVM decision value | 3.75 |
| SVM RNA-class probability | 0.999585 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18886779 120 + 27905053 AUCCGGUGGGGUGUCCUUCUUCAUGUACUUCCCCACCACGCCCUCCUUGUGCUGGGACUUCUCCUUGUUCCGGCGCGAGAGCAUCGUCGGUUUUCGGUUGCACAGCAGGAUCGUUAGAUA (((((((((((.((.(........).))..)))))))..((....(((((((((((((........))))))))))))).((((((........))).)))...)).))))......... ( -47.80) >DroSec_CAF1 1046 120 + 1 CUCCGGUGGGGUGUCCUUCUUCAUGUACUUCCCCACCACGCCCUCCUUGUGCUGGGACUUCUCCUUGUUCCGGCGCGAGAGCAUCGUCGGUUUUCGGUUGCACAGCAGGAUCGUUAGAUA .((((((((((.((.(........).))..)))))))..((....(((((((((((((........))))))))))))).((((((........))).)))...)).))).......... ( -47.30) >DroSim_CAF1 690 120 + 1 CUCCGGUGGGGUGUCCUUCUUCAUGUACUUCCCCACCACGCCCUCCUUGUGCUGGGACUUCUCCUUGUUCCGGCGCGAGAGCAUCGUCGGUUUUCGGUUGCACAGCAGGAUCGUUAGAUA .((((((((((.((.(........).))..)))))))..((....(((((((((((((........))))))))))))).((((((........))).)))...)).))).......... ( -47.30) >DroEre_CAF1 825 120 + 1 CUCCGGUGGGGUGUCCUUCUUCAUGUACUUCCCCACCACGCCCUCCUUGUGCUGGGACUUCUCCUUGUUCCGGCGCGAGAGCAUCGUCGGUUUUCGGUUGCACAGCAGGAUCGUUAGAUA .((((((((((.((.(........).))..)))))))..((....(((((((((((((........))))))))))))).((((((........))).)))...)).))).......... ( -47.30) >DroYak_CAF1 688 120 + 1 CUCCGGUGGGGUGUCCUUCUUCAUGUACUUCCCCACCACGCCCUCCUUGUGCUGGGACUUCUCCUUGUUCCGGCGCGAGAGCAUCGUCGGUUUUCGGUUGCACAGCAGGAUCGUUAGAUA .((((((((((.((.(........).))..)))))))..((....(((((((((((((........))))))))))))).((((((........))).)))...)).))).......... ( -47.30) >consensus CUCCGGUGGGGUGUCCUUCUUCAUGUACUUCCCCACCACGCCCUCCUUGUGCUGGGACUUCUCCUUGUUCCGGCGCGAGAGCAUCGUCGGUUUUCGGUUGCACAGCAGGAUCGUUAGAUA .((((((((((.((.(........).))..)))))))..((....(((((((((((((........))))))))))))).((((((........))).)))...)).))).......... (-47.34 = -47.34 + -0.00)

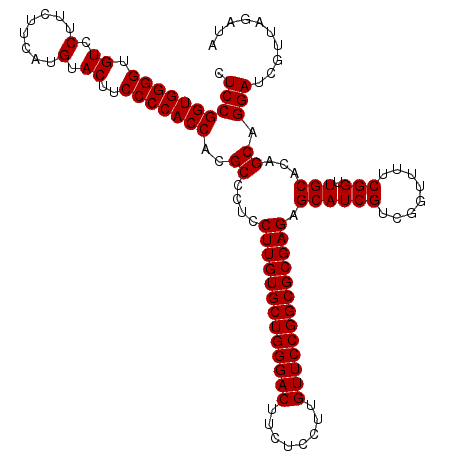
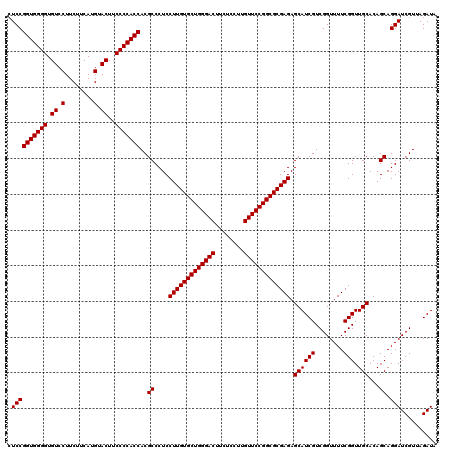
| Location | 18,886,819 – 18,886,939 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.33 |
| Mean single sequence MFE | -42.40 |
| Consensus MFE | -41.40 |
| Energy contribution | -41.44 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.779722 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18886819 120 + 27905053 CCCUCCUUGUGCUGGGACUUCUCCUUGUUCCGGCGCGAGAGCAUCGUCGGUUUUCGGUUGCACAGCAGGAUCGUUAGAUAGUUCAUGCAGACUAUGUUUCCGCUCGGAUUCCGGAUCGCU .....(((((((((((((........)))))))))))))(((....((((..(((((((((...)))(((.....(.((((((......)))))).).))))).))))..))))...))) ( -41.60) >DroSec_CAF1 1086 120 + 1 CCCUCCUUGUGCUGGGACUUCUCCUUGUUCCGGCGCGAGAGCAUCGUCGGUUUUCGGUUGCACAGCAGGAUCGUUAGAUAGUUCAUGCAGACUCUGUUUCCGCUCGGAUUCCGGAUCGUU .....(((((((((((((........)))))))))))))(((....((((..(((((..((..(((((..((((..((....))..)).))..)))))...)))))))..))))...))) ( -40.80) >DroSim_CAF1 730 120 + 1 CCCUCCUUGUGCUGGGACUUCUCCUUGUUCCGGCGCGAGAGCAUCGUCGGUUUUCGGUUGCACAGCAGGAUCGUUAGAUAGUUCAUGCAGACUCUGUUUCCGCUCGGAUUCCGGAUCGCU .....(((((((((((((........)))))))))))))(((....((((..(((((..((..(((((..((((..((....))..)).))..)))))...)))))))..))))...))) ( -43.20) >DroEre_CAF1 865 120 + 1 CCCUCCUUGUGCUGGGACUUCUCCUUGUUCCGGCGCGAGAGCAUCGUCGGUUUUCGGUUGCACAGCAGGAUCGUUAGAUAGUUCAUGCAGACUCUGUUUCCGCUCGGAUUCCGGAUCGCU .....(((((((((((((........)))))))))))))(((....((((..(((((..((..(((((..((((..((....))..)).))..)))))...)))))))..))))...))) ( -43.20) >DroYak_CAF1 728 120 + 1 CCCUCCUUGUGCUGGGACUUCUCCUUGUUCCGGCGCGAGAGCAUCGUCGGUUUUCGGUUGCACAGCAGGAUCGUUAGAUAGUUCAUGCAGACUCUGUUUCCGCUCGGAUUCCGGAUCGCU .....(((((((((((((........)))))))))))))(((....((((..(((((..((..(((((..((((..((....))..)).))..)))))...)))))))..))))...))) ( -43.20) >consensus CCCUCCUUGUGCUGGGACUUCUCCUUGUUCCGGCGCGAGAGCAUCGUCGGUUUUCGGUUGCACAGCAGGAUCGUUAGAUAGUUCAUGCAGACUCUGUUUCCGCUCGGAUUCCGGAUCGCU .....(((((((((((((........)))))))))))))(((....((((..(((((..((..(((((..((((..((....))..)).))..)))))...)))))))..))))...))) (-41.40 = -41.44 + 0.04)


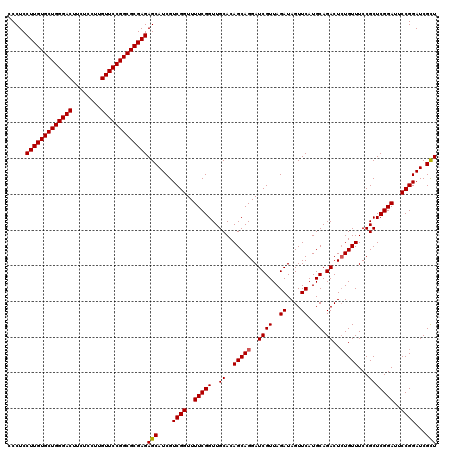
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:39:04 2006