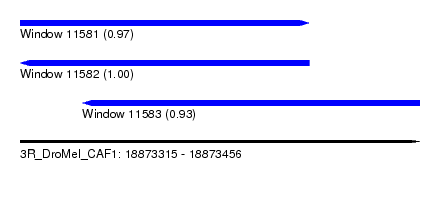
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,873,315 – 18,873,456 |
| Length | 141 |
| Max. P | 0.999934 |

| Location | 18,873,315 – 18,873,417 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.98 |
| Mean single sequence MFE | -24.00 |
| Consensus MFE | -20.54 |
| Energy contribution | -20.10 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.969662 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18873315 102 + 27905053 ----------------UAC--UUAACAUGAAAUGCGACUGCAUUUUCUAAGUGCAUCCAUGUCUGAUGUGCAUCUGACUAGCCUUAUUAAUGGCUCAUUUGCCAGACGCUCAUUGCAGCC ----------------.((--(((....(((((((....))))))).)))))(((....((((((....(((..(((...(((........))))))..))))))))).....))).... ( -25.60) >DroSim_CAF1 1101 86 + 1 ----------------------------------AGACUGCAUUUUCUAAGUGCAUCCAUGUCUGAUGUGCAUCUGACUAGCCUUAUUAAUGGCUCAUUUGCCAGACGCUCAUUGCAGCC ----------------------------------.((.((((((.....))))))))...(((((....(((..(((...(((........))))))..))))))))((.....)).... ( -21.70) >DroYak_CAF1 1138 120 + 1 CAUAAUCUAUUUUUGAUACGAUUAACGUGAAAGCAUACUGCAUUUUCCGAGUGCAUCCAAGUCCGAUGUGUAUCUGACUAGCCUUAUUAAUGGCUCAUUUGCCAGGCGUUCAUUGCAGCC ..(((((((((...)))).)))))..((....))...(((((......(((..((((.......))))..).))(((...((((.......(((......)))))))..))).))))).. ( -24.71) >consensus ________________UAC__UUAAC_UGAAA__AGACUGCAUUUUCUAAGUGCAUCCAUGUCUGAUGUGCAUCUGACUAGCCUUAUUAAUGGCUCAUUUGCCAGACGCUCAUUGCAGCC .....................................(((((........(..((((.......))))..).((((.(.((((........)))).....).)))).......))))).. (-20.54 = -20.10 + -0.44)

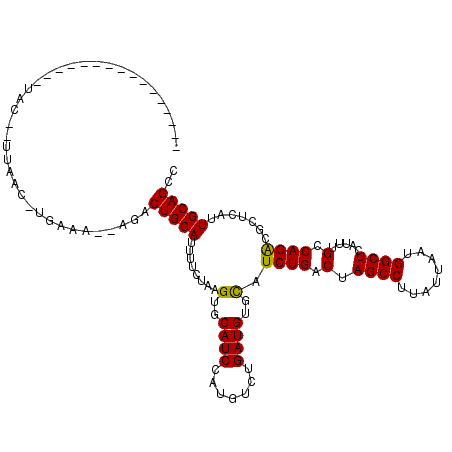
| Location | 18,873,315 – 18,873,417 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.98 |
| Mean single sequence MFE | -30.00 |
| Consensus MFE | -26.87 |
| Energy contribution | -28.20 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.90 |
| SVM decision value | 4.65 |
| SVM RNA-class probability | 0.999934 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18873315 102 - 27905053 GGCUGCAAUGAGCGUCUGGCAAAUGAGCCAUUAAUAAGGCUAGUCAGAUGCACAUCAGACAUGGAUGCACUUAGAAAAUGCAGUCGCAUUUCAUGUUAA--GUA---------------- (((((((....(((((((((.....((((........)))).))))))))).((((.......))))...........)))))))..............--...---------------- ( -33.20) >DroSim_CAF1 1101 86 - 1 GGCUGCAAUGAGCGUCUGGCAAAUGAGCCAUUAAUAAGGCUAGUCAGAUGCACAUCAGACAUGGAUGCACUUAGAAAAUGCAGUCU---------------------------------- (((((((....(((((((((.....((((........)))).))))))))).((((.......))))...........))))))).---------------------------------- ( -32.70) >DroYak_CAF1 1138 120 - 1 GGCUGCAAUGAACGCCUGGCAAAUGAGCCAUUAAUAAGGCUAGUCAGAUACACAUCGGACUUGGAUGCACUCGGAAAAUGCAGUAUGCUUUCACGUUAAUCGUAUCAAAAAUAGAUUAUG .((((((.....((....(((....((((........))))((((.(((....))).))))....)))...)).....))))))............(((((............))))).. ( -24.10) >consensus GGCUGCAAUGAGCGUCUGGCAAAUGAGCCAUUAAUAAGGCUAGUCAGAUGCACAUCAGACAUGGAUGCACUUAGAAAAUGCAGUCU__UUUCA_GUUAA__GUA________________ (((((((....(((((((((.....((((........)))).))))))))).((((.......))))...........)))))))................................... (-26.87 = -28.20 + 1.33)

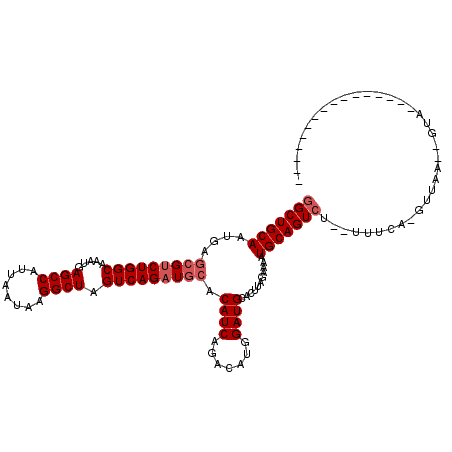
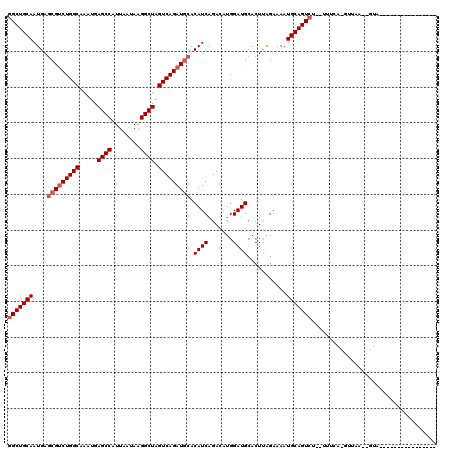
| Location | 18,873,337 – 18,873,456 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.20 |
| Mean single sequence MFE | -39.47 |
| Consensus MFE | -33.83 |
| Energy contribution | -35.17 |
| Covariance contribution | 1.34 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.93 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932362 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
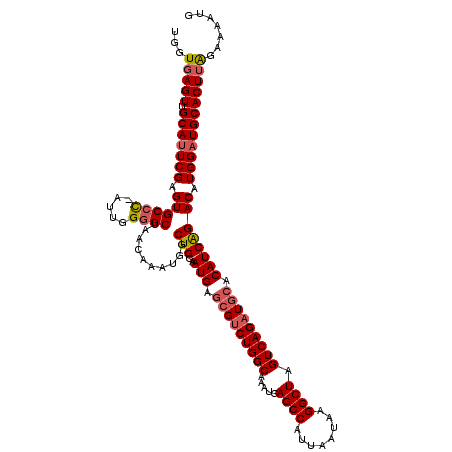
>3R_DroMel_CAF1 18873337 119 - 27905053 UGGUGAGUUGCAUUCGAGUGCCC-AUUGGGGCUAACAAAUGGCUGCAAUGAGCGUCUGGCAAAUGAGCCAUUAAUAAGGCUAGUCAGAUGCACAUCAGACAUGGAUGCACUUAGAAAAUG ...(((((.(((((((.((...(-((((.(((((.....))))).))))).(((((((((.....((((........)))).))))))))).......)).))))))))))))....... ( -45.00) >DroSim_CAF1 1107 120 - 1 UGGUGAGUUGCAUUCGAGUGCCCAAUUGGGGCUAACAAAUGGCUGCAAUGAGCGUCUGGCAAAUGAGCCAUUAAUAAGGCUAGUCAGAUGCACAUCAGACAUGGAUGCACUUAGAAAAUG ...(((((.((((((....((((.....))))......(((.(((..(((.(((((((((.....((((........)))).))))))))).)))))).))))))))))))))....... ( -43.80) >DroYak_CAF1 1178 119 - 1 UGGUAAGUUGCAAUCGAGUGCUU-AUUCGUGCUAACAAAUGGCUGCAAUGAACGCCUGGCAAAUGAGCCAUUAAUAAGGCUAGUCAGAUACACAUCGGACUUGGAUGCACUCGGAAAAUG .............((((((((..-((((((((((.....)))).)).......(((((((......))).......)))).((((.(((....))).)))).))))))))))))...... ( -29.61) >consensus UGGUGAGUUGCAUUCGAGUGCCC_AUUGGGGCUAACAAAUGGCUGCAAUGAGCGUCUGGCAAAUGAGCCAUUAAUAAGGCUAGUCAGAUGCACAUCAGACAUGGAUGCACUUAGAAAAUG ...(((((.(((((((.((((((.....))))..........(((..(((.(((((((((.....((((........)))).))))))))).)))))))).))))))))))))....... (-33.83 = -35.17 + 1.34)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:38:53 2006