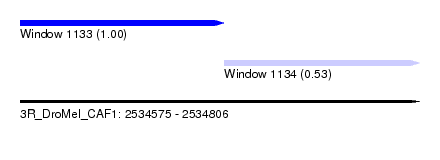
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,534,575 – 2,534,806 |
| Length | 231 |
| Max. P | 0.999215 |

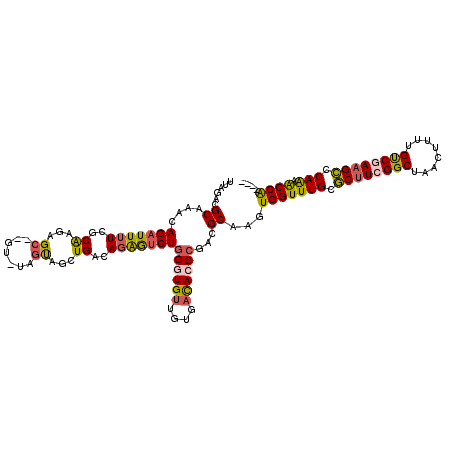
| Location | 2,534,575 – 2,534,693 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 73.26 |
| Mean single sequence MFE | -27.19 |
| Consensus MFE | -19.35 |
| Energy contribution | -18.80 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.71 |
| SVM decision value | 3.44 |
| SVM RNA-class probability | 0.999215 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2534575 118 + 27905053 UAUAAAGAUUUGCCUAAUGAGAAAAUUACAGUUUUAAAUGAGUGGCAUUUUUAGAUAAACCACUGAAUAGUUUACAUCAUUUUUGAAAAUUACGUUUUUCAAUGGGCAAAUAAUCGAC .......(((((((((.(((((((.......((..((((((((((....(((((........)))))....)))).))))))..))........))))))).)))))))))....... ( -26.66) >DroSec_CAF1 687 108 + 1 UCUAAAGAUUGGCCUGGUGCGAUAAUUUAAGCGUUUAAUGAGACGCAUGUUUG------CUAAUAAUUACUUUUAGCCAGUUA---UAACUGGG-CUUUAAAUGGGCCAAUAAUCGUC .......((((((((...((((.....((.((((((....)))))).)).)))------)..........((..(((((((..---..))).))-))..))..))))))))....... ( -27.00) >DroSim_CAF1 680 108 + 1 UAUAAAGAUUGGCCUGGUGCGAUAAUUACAGCGUUUAUUGAGACGCAUUUUUG------CUAAUAAUAACUUUUAGCCAGUUA---UAAUUGGG-CUUUAAAUGGGCCAAUAAUCGUC .......(((((((((((.((((.((.((.((((((....))))))......(------((((.........)))))..)).)---).)))).)-))......))))))))....... ( -27.90) >consensus UAUAAAGAUUGGCCUGGUGCGAUAAUUACAGCGUUUAAUGAGACGCAUUUUUG______CUAAUAAUUACUUUUAGCCAGUUA___UAAUUGGG_CUUUAAAUGGGCCAAUAAUCGUC .......(((((((((..............(((((......)))))..............................(((((((...))))))).........)))))))))....... (-19.35 = -18.80 + -0.55)

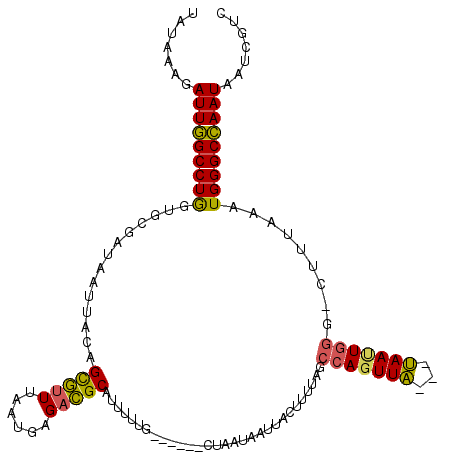
| Location | 2,534,693 – 2,534,806 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.45 |
| Mean single sequence MFE | -35.92 |
| Consensus MFE | -25.18 |
| Energy contribution | -25.10 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.70 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.530364 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2534693 113 + 27905053 UUAGACGCAAAAAGAUUAUCGCAGGAGC--AU-UAAUAGCUGACAGAAUCUGGGUGUUGUGACACCCUAAGCAAGUGGUUUGCUCUUCGGCUAACUUUUGUCGAAGACCAAAACCG---- ......((....(((((....(((....--..-......)))....)))))((((((....))))))...))....(((((..((((((((........))))))))...))))).---- ( -34.00) >DroSec_CAF1 795 113 + 1 UUAGACGCAAACAGAUUUUCGCAAGAGC--GU-UAGUAACUGACAGAGUCUGGAUGUUGUGAUACCCGACGCAAGUGGUUUGCGCUUCGGCUAAAUUUUGUCCAAGCCCAAAGCCA---- .....(((((.(((((((((....))..--((-(((...))))).)))))))....))))).......((....))(((((..((((.(((........))).))))...))))).---- ( -32.50) >DroSim_CAF1 788 113 + 1 UUAAACGCAAACAGAUUUUCGCAAGAGC--GU-UAGUAACUGACAGAGUCUGGAUGUUGUGAUACCCGACGCAAGUGGUUUGCGCUUCGGCUAACUUUUGUCCAAGCCCAAAACCA---- .....(((((.(((((((((....))..--((-(((...))))).)))))))....))))).......((....))(((((..((((.(((........))).))))...))))).---- ( -32.50) >DroEre_CAF1 1122 109 + 1 UUAGACGC----AGAUUUUCGCGAGGGC--GG-UUGCAGCUGACAGGAGCUAGGUGUUAUGUCACCCGGCGCAAGUGGUUUGUGCUUCGGCUAACGCAUGCCGAAGCUCAGAACCA---- ((((..((----((...(((....))).--..-))))..)))).....((..((((......))))..)).....((((((..((((((((........))))))))...))))))---- ( -41.90) >DroYak_CAF1 481 120 + 1 UAAGACGCAAUCAGAUUUUUGCAAGGGCAAGGUUAGUAGCUGACAGGGUCUGGGUGUUGUGACACCCCAUGCAAGUGGUUUGUGCCUCGGCUAACGCAUGUCGAAGCCCAGCACCAUAAC ..((((.(..((((..((((((....)))))).......))))..).))))((((((....)))))).......((((((((.((.(((((........))))).)).))).)))))... ( -38.70) >consensus UUAGACGCAAACAGAUUUUCGCAAGAGC__GU_UAGUAGCUGACAGAGUCUGGGUGUUGUGACACCCGACGCAAGUGGUUUGCGCUUCGGCUAACUUUUGUCGAAGCCCAAAACCA____ ......((....(((((((..((...((.......))...))..)))))))((((((....))))))...))...(((((((.((((((((........)))))))).))).)))).... (-25.18 = -25.10 + -0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:50:06 2006