| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,847,743 – 18,847,836 |
| Length | 93 |
| Max. P | 0.722757 |

| Location | 18,847,743 – 18,847,836 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 84.04 |
| Mean single sequence MFE | -21.72 |
| Consensus MFE | -18.26 |
| Energy contribution | -17.85 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.570962 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18847743 93 + 27905053 UCGAUGAUGACAAUGCAAUUUGACAACUCAUUAAAGUUCAUUGAAUUGUUAUUAGCUGCAACACCGG------------UGCCGCAAUUGCUGCGGUCUGUCGUC- .....(((((((.((((...((((((.(((...........))).)))))).....)))).......------------.((((((.....)))))).)))))))- ( -25.10) >DroPse_CAF1 93593 91 + 1 UCAAUGAUGACAAUGCAAUUUGACAACUCAUUAAAGUUCAUUGAAUUGUUAUUAGCUGCAACAU---------------UGCCGCAGACAUUGCGGUUUGUCCUGU ........(((((((((...((((((.(((...........))).)))))).....))))....---------------.(((((((...)))))))))))).... ( -21.00) >DroEre_CAF1 72674 93 + 1 UCGAUGAUGACAAUGCAAUUUGACAACUCAUUAAAGUUCAUUGAAUUGUUAUUAGCUGCAACACCGG------------UGCCGCAAUUGCUGCGGUCUGUCCUC- ....((((((((((.(((..(((..(((......))))))))).))))))))))..........(((------------.((((((.....))))))))).....- ( -21.20) >DroWil_CAF1 85050 89 + 1 UCAAUAAUGACAAUGCAAUUUGACAACUCAUUAAAGUUCAUUGAAUUGUUAUUAGCUGCAACAU---------------UGCCGCAGUUGUUGUGGUUAGCCAU-- ....((((.(((((((((((..(.((((......))))..)..))))))....((((((.....---------------....))))))))))).)))).....-- ( -19.70) >DroAna_CAF1 76618 102 + 1 UCAAUGAUGACAAUGCAAUUUGACAACUCAUUAAAGUUCAUUGAAUUGUUAUUAGCUGCAACACCACCCGUCGCAAUACGGCUGCAAUCAUUGCGGUCUAUC---- ....((((((((((.(((..(((..(((......))))))))).))))))))))..(((.((.......)).)))....(((((((.....)))))))....---- ( -22.30) >DroPer_CAF1 92739 91 + 1 UCAAUGAUGACAAUGCAAUUUGACAACUCAUUAAAGUUCAUUGAAUUGUUAUUAGCUGCAACAU---------------UGCCGCAGACAUUGCGGUUUGUCCUGU ........(((((((((...((((((.(((...........))).)))))).....))))....---------------.(((((((...)))))))))))).... ( -21.00) >consensus UCAAUGAUGACAAUGCAAUUUGACAACUCAUUAAAGUUCAUUGAAUUGUUAUUAGCUGCAACAC_______________UGCCGCAAUCAUUGCGGUCUGUCCU__ ....((((((((((.(((..(((..(((......))))))))).))))))))))..........................((((((.....))))))......... (-18.26 = -17.85 + -0.41)

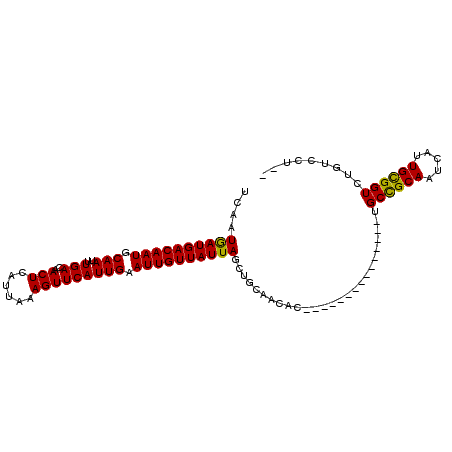

| Location | 18,847,743 – 18,847,836 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 84.04 |
| Mean single sequence MFE | -22.62 |
| Consensus MFE | -19.44 |
| Energy contribution | -18.81 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.722757 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18847743 93 - 27905053 -GACGACAGACCGCAGCAAUUGCGGCA------------CCGGUGUUGCAGCUAAUAACAAUUCAAUGAACUUUAAUGAGUUGUCAAAUUGCAUUGUCAUCAUCGA -((.(((((...((((((....((...------------.)).)))))).((.....((((((((.(((...))).))))))))......)).))))).))..... ( -24.10) >DroPse_CAF1 93593 91 - 1 ACAGGACAAACCGCAAUGUCUGCGGCA---------------AUGUUGCAGCUAAUAACAAUUCAAUGAACUUUAAUGAGUUGUCAAAUUGCAUUGUCAUCAUUGA ....(((((...((((((.(((((((.---------------..)))))))).....((((((((.(((...))).))))))))...))))).)))))........ ( -22.40) >DroEre_CAF1 72674 93 - 1 -GAGGACAGACCGCAGCAAUUGCGGCA------------CCGGUGUUGCAGCUAAUAACAAUUCAAUGAACUUUAAUGAGUUGUCAAAUUGCAUUGUCAUCAUCGA -((.(((((...((((((....((...------------.)).)))))).((.....((((((((.(((...))).))))))))......)).))))).))..... ( -24.10) >DroWil_CAF1 85050 89 - 1 --AUGGCUAACCACAACAACUGCGGCA---------------AUGUUGCAGCUAAUAACAAUUCAAUGAACUUUAAUGAGUUGUCAAAUUGCAUUGUCAUUAUUGA --.(((....)))...((((((((((.---------------..))))))).((((.(((((.((((.(((((....))))).....)))).))))).))))))). ( -17.20) >DroAna_CAF1 76618 102 - 1 ----GAUAGACCGCAAUGAUUGCAGCCGUAUUGCGACGGGUGGUGUUGCAGCUAAUAACAAUUCAAUGAACUUUAAUGAGUUGUCAAAUUGCAUUGUCAUCAUUGA ----.........(((((((.((((......(((((((.....)))))))((.....((((((((.(((...))).))))))))......)).)))).))))))). ( -25.50) >DroPer_CAF1 92739 91 - 1 ACAGGACAAACCGCAAUGUCUGCGGCA---------------AUGUUGCAGCUAAUAACAAUUCAAUGAACUUUAAUGAGUUGUCAAAUUGCAUUGUCAUCAUUGA ....(((((...((((((.(((((((.---------------..)))))))).....((((((((.(((...))).))))))))...))))).)))))........ ( -22.40) >consensus __AGGACAAACCGCAACAACUGCGGCA_______________AUGUUGCAGCUAAUAACAAUUCAAUGAACUUUAAUGAGUUGUCAAAUUGCAUUGUCAUCAUUGA ....(((((...((((...((((((((................))))))))......((((((((.(((...))).))))))))....)))).)))))........ (-19.44 = -18.81 + -0.64)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:38:43 2006