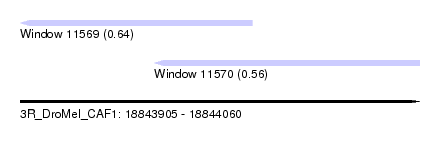
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,843,905 – 18,844,060 |
| Length | 155 |
| Max. P | 0.638059 |

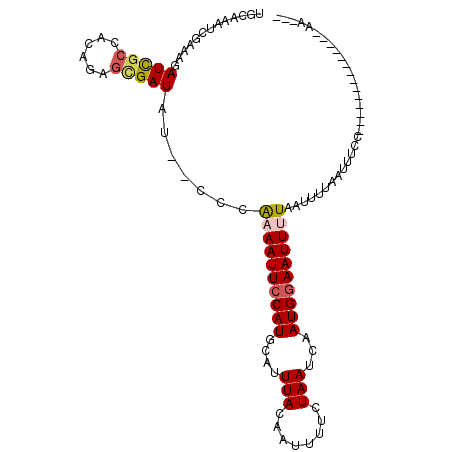
| Location | 18,843,905 – 18,843,995 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 82.15 |
| Mean single sequence MFE | -14.30 |
| Consensus MFE | -9.77 |
| Energy contribution | -10.13 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.638059 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18843905 90 - 27905053 UGCAAAUCGAAAGAUCGCCACACAGCGAUAU--CCUAAAAUUCCAUGCAUUUACAAUUUUCUAAUCAAUGGAAUUAUAAUUUUAAUUUCC--------------AA--- ((((..((....))((((......))))...--............))))...................((((((((......))).))))--------------).--- ( -14.80) >DroPse_CAF1 89498 90 - 1 UGCAAAUCCAAAGAUCACCACAGAGCGAUAU--ACCAAAAUUCCAUGCAUUUACAAUUUUCUAAUCAAUGGAAUUUUAAUUUUAAUUUCC--------------AA--- .............(((.(......).)))..--...((((((((((....(((........)))...)))))))))).............--------------..--- ( -8.70) >DroGri_CAF1 76353 104 - 1 UGCAAAUCGGCAGAUUGCCACAUCAUGAUAC--GUUGAAAUUCCAUGCAUUUACAAUUUUCUAAUCGAUGAAAUUUUAAUUUAUGUCGACACUGUGGAUUUUCGAA--- ........(((.....)))............--.((((((.((((((......)..........((((((((((....)))).))))))....))))).)))))).--- ( -17.20) >DroWil_CAF1 74485 95 - 1 UGCAAAUCAAAAGAUCGCCACAGAGCGAUAUACCCCAAAAUUCCAUGCAUUUAAAAUUUUCUAAUCAAUGGAAUUUUAAUUUUAAUUUAU--------------AGUGU ...((((.((((.(((((......))))).......((((((((((....(((........)))...))))))))))..)))).))))..--------------..... ( -14.80) >DroYak_CAF1 73821 90 - 1 UGCAAAUCGAAAGAUCGCCACACAGCGAUAU--CCGGAAAUUCCAUGCAUUUACAAUUUUCUAAUCAAUGGAAUUAUAAUUUUAAUUUCC--------------AA--- ((((..((....))((((......))))...--..((.....)).))))...................((((((((......))).))))--------------).--- ( -16.20) >DroPer_CAF1 88613 90 - 1 UGCAAAUCCAAAGAUCGCCACAGAGCGAUAU--ACCAAAAUUCCAUGCAUUUACAAUUUUCUAAUCAAUGGAAUUUUAAUUUUAAUUUCC--------------AA--- .............(((((......)))))..--...((((((((((....(((........)))...)))))))))).............--------------..--- ( -14.10) >consensus UGCAAAUCGAAAGAUCGCCACAGAGCGAUAU__CCCAAAAUUCCAUGCAUUUACAAUUUUCUAAUCAAUGGAAUUUUAAUUUUAAUUUCC______________AA___ .............(((((......))))).......((((((((((....(((........)))...))))))))))................................ ( -9.77 = -10.13 + 0.36)

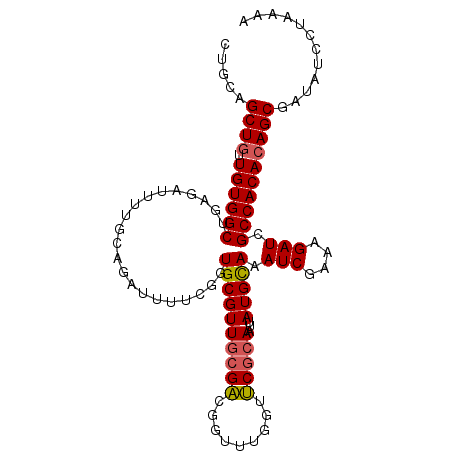
| Location | 18,843,957 – 18,844,060 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 89.13 |
| Mean single sequence MFE | -32.48 |
| Consensus MFE | -26.01 |
| Energy contribution | -26.10 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.557696 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18843957 103 - 27905053 CUGCUGCUGUUGUGGCUGAGAUUUUGCAGAUUUUCGGUGCGUUGCGACGGUUUGGUUCGCAAUUAUGCAAAUCGAAAGAUCGCCACACAGCGAUAUCCUAAAA ....((((((.(((((...((((((...(((((.((....((((((((......).)))))))..)).)))))..)))))))))))))))))........... ( -32.70) >DroSec_CAF1 68834 103 - 1 CUGCAGCUGUUGUGGCUGAGAUUUUGCAGAUUUUCGGUGCGUUGCGACGCUUUGGUUCGCAAUUAUGUAAAUCGAAAGAUCGCCACACAGCGAUAUCCUAAAA .....(((((.(((((...((((((...(((((.((....(((((((.((....)))))))))..)).)))))..))))))))))))))))............ ( -33.70) >DroSim_CAF1 72304 103 - 1 CUGCAGCUGUUGUGGCUGAGAUUUUGCAGAUUUUCGGUGCGUUGCGACGCUUUGGUUCGCAAUUAUGCAAAUCGAAAGAUCGCCACACAGCGAUAUCCUAAAA .....(((((.(((((...((((((...(((((.((....(((((((.((....)))))))))..)).)))))..))))))))))))))))............ ( -33.70) >DroYak_CAF1 73873 103 - 1 CUGCAGCUGUUGUGGCUGAGAUUUUGCAGAUUUUCGGUGCGUUGCGACGGUUUGGUUCGCAAUUAUGCAAAUCGAAAGAUCGCCACACAGCGAUAUCCGGAAA .....(((((.(((((...((((((...(((((.((....((((((((......).)))))))..)).)))))..))))))))))))))))............ ( -32.80) >DroAna_CAF1 73160 94 - 1 CUGCAGCUGUUGUGGCU---------CAGAUUUUCGGUGCGUUGGGACGGUUUGGUUCACAAUUAUGCAAAUCGAAAGAUCGCCACACAGCGAUAUCCUAAAA .....(((((.(((((.---------..((((((((((((((((((((......)))).))...)))))..))).))))))))))))))))............ ( -29.70) >DroPer_CAF1 88665 94 - 1 CUGCUGCUGCUGUGGCU---------CCGAUUUUCAGUGCGUUUCGGCGGUUUGUUCCGCAAUUAUGCAAAUCCAAAGAUCGCCACAGAGCGAUAUACCAAAA ....((((.(((((((.---------..((((((...(((((....((((......))))....)))))......)))))))))))))))))........... ( -32.30) >consensus CUGCAGCUGUUGUGGCUGAGAUUUUGCAGAUUUUCGGUGCGUUGCGACGGUUUGGUUCGCAAUUAUGCAAAUCGAAAGAUCGCCACACAGCGAUAUCCUAAAA .....((((.((((((.....................((((((((((.........)))))...))))).(((....))).))))))))))............ (-26.01 = -26.10 + 0.09)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:38:41 2006