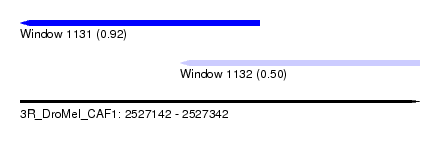
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,527,142 – 2,527,342 |
| Length | 200 |
| Max. P | 0.919199 |

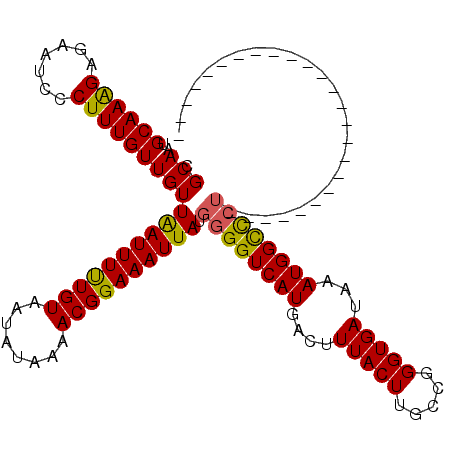
| Location | 2,527,142 – 2,527,262 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.24 |
| Mean single sequence MFE | -26.58 |
| Consensus MFE | -23.56 |
| Energy contribution | -23.08 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.919199 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2527142 120 - 27905053 CGCAAUGCAAAGAGAAUCCCUUUGUUGUUAAUUUUUGUAAUAUAAAACGGAAAUUAUGGGGUCAUGACUUUACUUGCCGGGUGAUAAAUGGCCGGGUGAUAAAAUGGGCCAUCCCGGCCA .(((..((((((.......)))))))))((((((((((........))))))))))((((((....))))))...((((((......((((((.(.........).)))))))))))).. ( -33.00) >DroSec_CAF1 55780 95 - 1 CGCAAUGCAAAGAGAAUCCCUUUGUUGUUAAUUUUUGUAAUAUAAAACGGAAAUUAUGGGGUCAUGACUUUACUUGCCGGGUGAUAAAUGGCCCU------------------------- .(((..((((((.......)))))))))((((((((((........)))))))))).((((((((....(((((.....)))))...))))))))------------------------- ( -25.40) >DroSim_CAF1 54714 95 - 1 CGCAAUGCAAAGAGAAUCCCUUUGUUGUUAAUUUUUGUAAUAUAAAACGGAAAUUAUGGGGUCAUGACUUUACUUGCCGGGUGAUAAAUGGCCCU------------------------- .(((..((((((.......)))))))))((((((((((........)))))))))).((((((((....(((((.....)))))...))))))))------------------------- ( -25.40) >DroEre_CAF1 55576 95 - 1 CGCAAUGCAAGGAGAAUCCCUUUGUUGUUGAUUUUUGUAAUAUAAAACGGAAAUUAUGGGGUCAUGACUUUACUUGCCGGGUGAUAAAUGGCUCU------------------------- .((((((.((((......))))))))))((((((((((........)))))))))).((((((((....(((((.....)))))...))))))))------------------------- ( -24.10) >DroYak_CAF1 57098 95 - 1 CGCAAUGCAAAGAGAAUCCCUUUGUUGUUGAUUUCUGUAAUAUAAAACGGAAAUUAUGGGGUCAUGACUUUACUUGUCGGGUGAUAAAUGGUCCU------------------------- .(((..((((((.......)))))))))((((((((((........)))))))))).(((..(((....(((((.....)))))...)))..)))------------------------- ( -25.00) >consensus CGCAAUGCAAAGAGAAUCCCUUUGUUGUUAAUUUUUGUAAUAUAAAACGGAAAUUAUGGGGUCAUGACUUUACUUGCCGGGUGAUAAAUGGCCCU_________________________ .(((..((((((.......)))))))))((((((((((........)))))))))).((((((((....(((((.....)))))...))))))))......................... (-23.56 = -23.08 + -0.48)

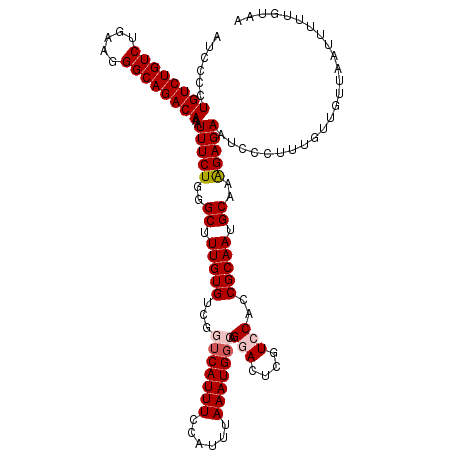
| Location | 2,527,222 – 2,527,342 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.33 |
| Mean single sequence MFE | -32.84 |
| Consensus MFE | -27.22 |
| Energy contribution | -28.06 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.83 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2527222 120 - 27905053 AUCCCCUGUCUGUCUGAAGGGCAGACAAAUUUCUGGGCUUUGUGUCGGUCAUUUCCAUUUAAAUGGCAGGACUCGUCCACCGCAAUGCAAAGAGAAUCCCUUUGUUGUUAAUUUUUGUAA ......((((((((.....))))))))..(((((..((.(((((...(((((((......))))))).(((....)))..))))).))..)))))......................... ( -32.80) >DroSec_CAF1 55835 120 - 1 AUCCCCUGUCUGUCUGAAGGGCAGACAAAUUUCUGGGCUUUGUGUCGGUCAUUUCCAUUUAAAUGGCAGGACUCGUGCACCGCAAUGCAAAGAGAAUCCCUUUGUUGUUAAUUUUUGUAA ......((((((((.....)))))))).......((((..((.(((.(((((((......)))))))..))).)).)).))(((..((((((.......)))))))))............ ( -31.30) >DroSim_CAF1 54769 120 - 1 AUCCCCUGUCUGUCUGAAGGGCAGACAAAUUUCUGAGCUUUGUGUCGGUCAUUUCCAUUUAAAUGGCAGGACUCGUCCACCGCAAUGCAAAGAGAAUCCCUUUGUUGUUAAUUUUUGUAA ......((((((((.....))))))))..(((((..((.(((((...(((((((......))))))).(((....)))..))))).))..)))))......................... ( -31.80) >DroEre_CAF1 55631 120 - 1 AUGCCCUGUCUGUCUGAAGGGCAGACAAAUUUCUGGGCUUUGUGUCGGUCAUUUCCAUUUAAAUGCAAGGACUCGUCCACCGCAAUGCAAGGAGAAUCCCUUUGUUGUUGAUUUUUGUAA ..((((((((((((.....)))))))).......))))........((......)).......((((((((....))).(.((((((.((((......)))))))))).)....))))). ( -32.11) >DroYak_CAF1 57153 120 - 1 AUCCUCUGUCUGUCUGAAGGGCAGACAAAUUUCUGGGCUUUGUGGCGGUCAUUUCCAUUCAAAUGGAAGAAAUCGUCCACCGCAAUGCAAAGAGAAUCCCUUUGUUGUUGAUUUCUGUAA ......((((((((.....))))))))..(((((..((.((((((.((.(..((((((....))))))(....)).)).)))))).))..)))))......................... ( -36.20) >consensus AUCCCCUGUCUGUCUGAAGGGCAGACAAAUUUCUGGGCUUUGUGUCGGUCAUUUCCAUUUAAAUGGCAGGACUCGUCCACCGCAAUGCAAAGAGAAUCCCUUUGUUGUUAAUUUUUGUAA ......((((((((.....))))))))..(((((..((.(((((...(((((((......))))))).(((....)))..))))).))..)))))......................... (-27.22 = -28.06 + 0.84)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:50:05 2006