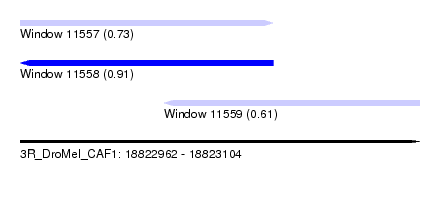
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,822,962 – 18,823,104 |
| Length | 142 |
| Max. P | 0.905840 |

| Location | 18,822,962 – 18,823,052 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 91.10 |
| Mean single sequence MFE | -35.92 |
| Consensus MFE | -26.06 |
| Energy contribution | -26.22 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.732063 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18822962 90 + 27905053 GUGCUGAAG--------UGCAUAUGUGCAUAUCUGAGCACGCAUUGUGCACUU-------GGCUGUCGGAGCCACUUCGCUUGGCUGGCCAACAAAGCGUUGGCA ..(((.(((--------(((((((((((........)))))...)))))))))-------)))......(((((.......))))).((((((.....)))))). ( -35.20) >DroSec_CAF1 48203 90 + 1 GUGCUGAAG--------UGCAUAUGCGCAUAUCUGAGCACGCAUUGUGCACUU-------GGCUGUCGGAGCCACUUCGCUUGGCUGGCCAACAAAGCGUUGGCA ..(((.(((--------((((((((((............)))).)))))))))-------)))......(((((.......))))).((((((.....)))))). ( -34.00) >DroSim_CAF1 51381 90 + 1 GUGCUGAAG--------UGCAUAUGCGCAUAUCUGAGCACGCAUUGUGCACUU-------GGCUGUCGGAGCCACUUCGCUUGGCUGGCCAACAAAGCGUUGGCA ..(((.(((--------((((((((((............)))).)))))))))-------)))......(((((.......))))).((((((.....)))))). ( -34.00) >DroEre_CAF1 48374 89 + 1 GUGCUG---------UGUGCAUAUGUGCAUAUCUGAGCACGCAUUGUGCACUU-------GGCUGUCGGAGCCACUUCGCUUGGCUGGCCAACAAAGCGUUGGCA ((((((---------((((((....)))))))...)))))((...(((....(-------((((.....)))))...)))...))..((((((.....)))))). ( -34.40) >DroYak_CAF1 51600 105 + 1 GUGCUGAAGUGCAUAUGUGCAAAUGUGCAUAUCUGAGCACGCAUUGUGCACUUGGCUCUUGGCUGUCGGAGCCACUUCGCUUGGCUGGCCAACAAAGCGUUGGCA (((((((.((((((((......)))))))).))..)))))((...(((....(((((((........)))))))...)))...))..((((((.....)))))). ( -42.00) >consensus GUGCUGAAG________UGCAUAUGUGCAUAUCUGAGCACGCAUUGUGCACUU_______GGCUGUCGGAGCCACUUCGCUUGGCUGGCCAACAAAGCGUUGGCA ..(((((..........((((((((((............)))).))))))..........((((.....)))).......)))))..((((((.....)))))). (-26.06 = -26.22 + 0.16)

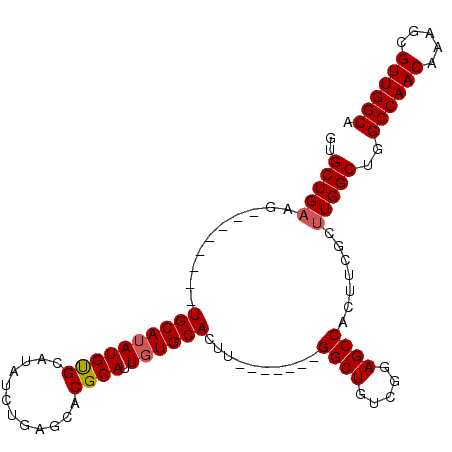
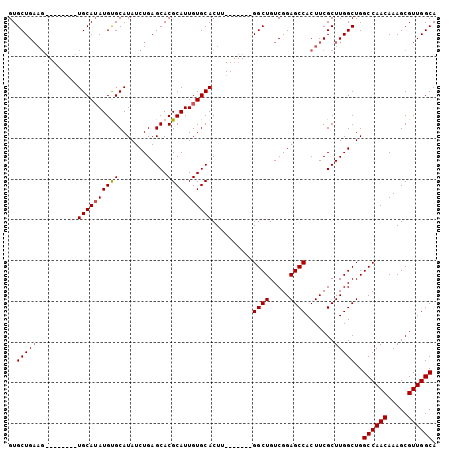
| Location | 18,822,962 – 18,823,052 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 91.10 |
| Mean single sequence MFE | -31.04 |
| Consensus MFE | -24.58 |
| Energy contribution | -24.34 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.905840 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18822962 90 - 27905053 UGCCAACGCUUUGUUGGCCAGCCAAGCGAAGUGGCUCCGACAGCC-------AAGUGCACAAUGCGUGCUCAGAUAUGCACAUAUGCA--------CUUCAGCAC (((....((((((((((....)).))))))))((((.....))))-------(((((((...((((((......))))))....))))--------)))..))). ( -29.30) >DroSec_CAF1 48203 90 - 1 UGCCAACGCUUUGUUGGCCAGCCAAGCGAAGUGGCUCCGACAGCC-------AAGUGCACAAUGCGUGCUCAGAUAUGCGCAUAUGCA--------CUUCAGCAC (((....((((((((((....)).))))))))((((.....))))-------(((((((..((((((..........)))))).))))--------)))..))). ( -31.50) >DroSim_CAF1 51381 90 - 1 UGCCAACGCUUUGUUGGCCAGCCAAGCGAAGUGGCUCCGACAGCC-------AAGUGCACAAUGCGUGCUCAGAUAUGCGCAUAUGCA--------CUUCAGCAC (((....((((((((((....)).))))))))((((.....))))-------(((((((..((((((..........)))))).))))--------)))..))). ( -31.50) >DroEre_CAF1 48374 89 - 1 UGCCAACGCUUUGUUGGCCAGCCAAGCGAAGUGGCUCCGACAGCC-------AAGUGCACAAUGCGUGCUCAGAUAUGCACAUAUGCACA---------CAGCAC .((((((.....)))))).......((....(((((.....))))-------).(((((...((((((......))))))....))))).---------..)).. ( -28.30) >DroYak_CAF1 51600 105 - 1 UGCCAACGCUUUGUUGGCCAGCCAAGCGAAGUGGCUCCGACAGCCAAGAGCCAAGUGCACAAUGCGUGCUCAGAUAUGCACAUUUGCACAUAUGCACUUCAGCAC .((((((.....)))))).......((((((((((((..........)))))..(((((.(((..((((........))))))))))))......))))).)).. ( -34.60) >consensus UGCCAACGCUUUGUUGGCCAGCCAAGCGAAGUGGCUCCGACAGCC_______AAGUGCACAAUGCGUGCUCAGAUAUGCACAUAUGCA________CUUCAGCAC .......(((.((((((..(((((.......)))))))))))((............((.....))((((........))))....)).............))).. (-24.58 = -24.34 + -0.24)

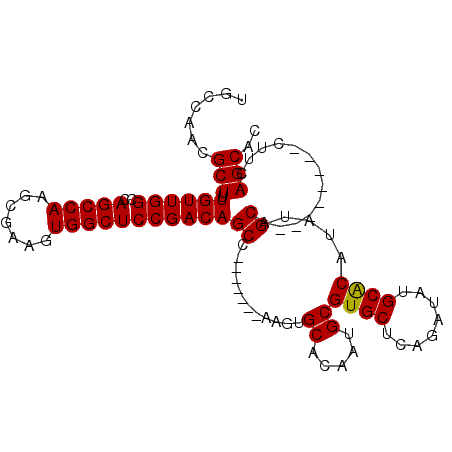
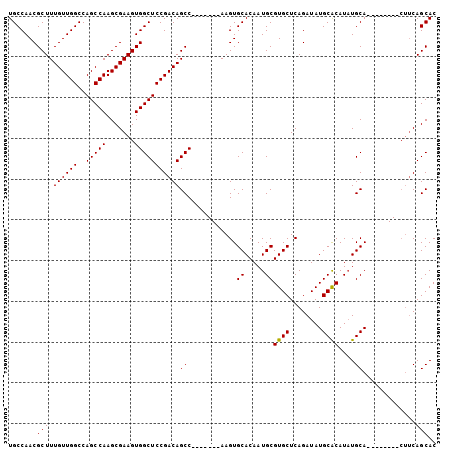
| Location | 18,823,013 – 18,823,104 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 78.11 |
| Mean single sequence MFE | -31.92 |
| Consensus MFE | -14.80 |
| Energy contribution | -14.36 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.608712 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
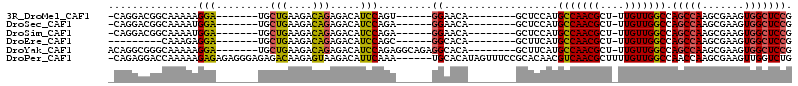
>3R_DroMel_CAF1 18823013 91 - 27905053 -CAGGACGGCAAAAAGGA-------UGCUGAAGACAGAGACAUCCAGU------GGAACA--------GCUCCAUGCCAACGCU-UUGUUGGCCAGCCAAGCGAAGUGGCUCCG -..(((((((.....(((-------((((........)).))))).((------(((...--------..))))))))..((((-(((((((....)).)))))))))).))). ( -34.00) >DroSec_CAF1 48254 91 - 1 -CAGGACGGCAAAAUGGA-------UGCUGAAGACAGAGACAUCCAGA------GGAACA--------GCUCCAUGCCAACGCU-UUGUUGGCCAGCCAAGCGAAGUGGCUCCG -..((((((((...((((-------((((........)).))))))..------(((...--------..))).))))..((((-(((((((....)).)))))))))).))). ( -33.70) >DroSim_CAF1 51432 91 - 1 -CAGGACGGCAAAAUGGA-------UGCUGAAGACAGAGACAUCCAGA------GGAACA--------GCUCCAUGCCAACGCU-UUGUUGGCCAGCCAAGCGAAGUGGCUCCG -..((((((((...((((-------((((........)).))))))..------(((...--------..))).))))..((((-(((((((....)).)))))))))).))). ( -33.70) >DroEre_CAF1 48424 83 - 1 ---------CAAAGAGGA-------UGCUGAAGACAGAGACAUCCAGC------GGCACA--------GCUUCAUGCCAACGCU-UUGUUGGCCAGCCAAGCGAAGUGGCUCCG ---------......(((-------((((........)).)))))(((------((((..--------......))))..((((-(((((((....)).))))))))))))... ( -28.30) >DroYak_CAF1 51666 98 - 1 ACAGGCGGGCAAAAAGGA-------UGCUGAAGACAGAGACAUCCAGAGGCAGAGGCACA--------GCUUCAUGCCAACGCU-UUGUUGGCCAGCCAAGCGAAGUGGCUCCG .....(((((.....(((-------((((........)).)))))...(((((((((...--------))))).))))..((((-(((((((....)).))))))))))).))) ( -37.80) >DroPer_CAF1 62832 107 - 1 -CAGAGGACCAAAAAGAGAGAGGGAGAGACAAGAGUAAGACAUUCAAA------UGCACAUAGUUUCCGCACAACGUCAACGCUUUUGUUGGCCAACCAAGCGAAGUUGGUCUG -....(((((((...(......((...(((((((((..(.(((....)------))).....(((..((.....))..))))))))))))..)).......)....))))))). ( -24.02) >consensus _CAGGACGGCAAAAAGGA_______UGCUGAAGACAGAGACAUCCAGA______GGAACA________GCUCCAUGCCAACGCU_UUGUUGGCCAGCCAAGCGAAGUGGCUCCG ...............(((.........(((....))).....))).........((...................(((((((....))))))).(((((.......))))))). (-14.80 = -14.36 + -0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:38:31 2006