| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
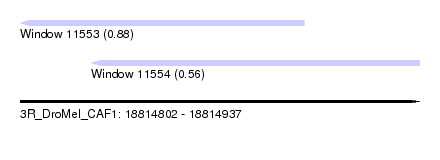
| Location | 18,814,802 – 18,814,937 |
| Length | 135 |
| Max. P | 0.876787 |

| Location | 18,814,802 – 18,814,898 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 71.14 |
| Mean single sequence MFE | -20.52 |
| Consensus MFE | -12.60 |
| Energy contribution | -13.43 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.876787 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18814802 96 - 27905053 GGUGAACUAGGGUGGUAUCAUUUUACUUUGUCCAGCUUGAA-CUUACUUUCUACAUCCUAGUUCAC--UUCUAGUUCCUUAAAAUUACUGCUGA--CAUCC--- (((((((((((((((((......)))............(((-......)))..)))))))))))))--).........................--.....--- ( -22.20) >DroSec_CAF1 40032 96 - 1 GGUGAACUAGGGUGGUAUCAUUUUACUUAGUCCAGCUUGAA-CUUACUUUCUACAUCCUAGUUCGC--UCCUAGUUCCUUAAAAUUACUACUGA--CAUCC--- (((((((((((((((((......)))............(((-......)))..)))))))))))))--).........................--.....--- ( -21.80) >DroSim_CAF1 43203 96 - 1 GGUGAACUAGGGUGGUAUCAUUUUACUAAGUCCAGCUUGAA-CUUACUUUCUACAUCCUAGUUCAC--UCCUAGUUCCUUAAAAUUACUACUGA--CAUCC--- (((((((((((((((((......)))(((((.(.....).)-)))).......)))))))))))))--).........................--.....--- ( -24.60) >DroEre_CAF1 40291 94 - 1 GGUGAACUAGGGUGGUAUCAAUAAACUUUUCCCUCGCUGAA-CUCUCUUUUUACGUCCUAGUUCAC--UCCAAGCGCC-UGGACAUUCUGCUGG--CAUC---- (((((((((((..(...........................-...........)..))))))))))--)......(((-.((.....))...))--)...---- ( -21.76) >DroYak_CAF1 42736 84 - 1 UGUGAACUAGGGCGGUAUCAAUAGACUUUCUCCUGGCUGAA-CCUACAUUUUACAUCCUAGUUC------------CU-UGAAAUUACUGCUGG--CAUC---- ...(((((((((.(((.(((.(((........)))..))))-))...........)))))))))------------..-...............--....---- ( -14.20) >DroAna_CAF1 42482 101 - 1 CCCGAACUAGGGUAGUAUUGUCAUACUG-GUUCACGUUGAAACUAGCUUCUAAGAUUCCAGUUCAUGGUAAUAGUUA--AAAAUUUGCUUCUGAAGCAGCCUGC (((......)))...((((..(((((((-(........(((......))).......)))))..)))..))))....--.....(((((.....)))))..... ( -18.56) >consensus GGUGAACUAGGGUGGUAUCAUUAUACUUUGUCCAGCUUGAA_CUUACUUUCUACAUCCUAGUUCAC__UCCUAGUUCC_UAAAAUUACUGCUGA__CAUCC___ .(((((((((((((.......................................)))))))))))))...................................... (-12.60 = -13.43 + 0.83)

| Location | 18,814,826 – 18,814,937 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 70.98 |
| Mean single sequence MFE | -26.64 |
| Consensus MFE | -13.29 |
| Energy contribution | -13.88 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.560455 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18814826 111 - 27905053 CCUCAAUGGGUAGCCUUGUGGAAUUUU-UUGUUGAUUCAAGGUGAACUAGGGUGGUAUCAUUUUACUUUGUCCAGCUUGAA-CUUACUUUCUACAUCCUAGUUCAC--UUCUAGU ..(((((..(...((....)).....)-..)))))....((((((((((((((((((......)))............(((-......)))..)))))))))))))--))..... ( -29.20) >DroSec_CAF1 40056 112 - 1 CCCUAAUGGGCACCCUUGUGGAAUUUUUUUUUUGAUUCAAGGUGAACUAGGGUGGUAUCAUUUUACUUAGUCCAGCUUGAA-CUUACUUUCUACAUCCUAGUUCGC--UCCUAGU (((....)))(((....)))(((((........)))))..(((((((((((((((((......)))............(((-......)))..)))))))))))))--)...... ( -29.10) >DroSim_CAF1 43227 112 - 1 CCUUAAUGGGCAGCCUUGUGGAAUUUUUUUUUUGAUUCAAGGUGAACUAGGGUGGUAUCAUUUUACUAAGUCCAGCUUGAA-CUUACUUUCUACAUCCUAGUUCAC--UCCUAGU ......((((..........(((((........)))))..(((((((((((((((((......)))(((((.(.....).)-)))).......)))))))))))))--))))).. ( -30.40) >DroEre_CAF1 40313 111 - 1 CCUUAUUGGGUAAGUUAGUGGACAUUU-UUUUGGAUUUUGGGUGAACUAGGGUGGUAUCAAUAAACUUUUCCCUCGCUGAA-CUCUCUUUUUACGUCCUAGUUCAC--UCCAAGC (((....)))...(((....)))....-........(((((((((((((((..(...........................-...........)..))))))))))--.))))). ( -23.66) >DroYak_CAF1 42757 101 - 1 CCUUUAUGGGUGACCUUGUGGAAAUUU-UUUU-GAUUUAGUGUGAACUAGGGCGGUAUCAAUAGACUUUCUCCUGGCUGAA-CCUACAUUUUACAUCCUAGUUC----------- ((.....((....))....))......-....-..........(((((((((.(((.(((.(((........)))..))))-))...........)))))))))----------- ( -19.40) >DroAna_CAF1 42509 114 - 1 UAUUUAUAGGGGUUUCCCUGGGAUCUUUCUGUUAAUAAAGCCCGAACUAGGGUAGUAUUGUCAUACUG-GUUCACGUUGAAACUAGCUUCUAAGAUUCCAGUUCAUGGUAAUAGU .......((((....))))((((((((..((.(((((..((((......))))..))))).))..(((-((((.....).)))))).....))))))))................ ( -28.10) >consensus CCUUAAUGGGUAACCUUGUGGAAUUUU_UUUUUGAUUCAAGGUGAACUAGGGUGGUAUCAUUAUACUUUGUCCAGCUUGAA_CUUACUUUCUACAUCCUAGUUCAC__UCCUAGU (((....)))...............................(((((((((((((.......................................)))))))))))))......... (-13.29 = -13.88 + 0.59)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:38:26 2006