
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,806,480 – 18,806,583 |
| Length | 103 |
| Max. P | 0.923069 |

| Location | 18,806,480 – 18,806,572 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 79.53 |
| Mean single sequence MFE | -17.60 |
| Consensus MFE | -11.07 |
| Energy contribution | -11.27 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.923069 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18806480 92 + 27905053 ---GGACAA--C-UCGAUGCUGGUUGGCCCCACAAUCACUUUUCAAUUAAAUGGCAUUCUUUCCUCUUUGCCAUUAUUUUGUGUUUUUUUUUUGCUUU ---(((((.--.-..((...((((((......))))))....)).....(((((((............)))))))......)))))............ ( -15.40) >DroSec_CAF1 31871 84 + 1 ---GGACAA--CUCCGAUGCUGGUUGGCCCCACAAUCACUUUUCAAUUAAAUGGCAUUCUUUCCUCUUUGCCAUUAUUUUGUGUUUUUU--------- ---(((...--.)))(((((((((((......))))))...........(((((((............))))))).....)))))....--------- ( -16.30) >DroSim_CAF1 34406 78 + 1 ---UG--------UCGAUGCUGGUUGGCCCCACAAUCACUUUUCAAUUAAAUGGCAUUCUUUCCUCUUUGCCAUUAUUUUGUGUUUUUU--------- ---..--------..(((((((((((......))))))...........(((((((............))))))).....)))))....--------- ( -14.30) >DroEre_CAF1 32407 84 + 1 UGGGGGCAA----------CUGGUUGGCCCCACAAUCACUUUUCAAUUAAAUGGCAUUCUUUCCUCUUUGCCAUUAUUUUGUGUUUUUU----GCUCU (((((.(((----------....))).)))))(((..((....(((...(((((((............)))))))...))).))...))----).... ( -21.80) >DroYak_CAF1 34170 94 + 1 AGGGGGCAGAAGCUCGAUGUUGGUUGGCCCCACAAUCACUUUUCAAUUAAAUGGCAUUCUUUCCUCUUUGCCAUUAUUUUGUGUUUUUU----GCUCC ..(((((((((((.(((...((((((......))))))...........(((((((............)))))))...))).).)))))----))))) ( -26.20) >DroAna_CAF1 33334 89 + 1 UCGGAAAAA---CUCGAUUCUGGUUGGCCCCACAAUCACUUUUCAAUUAAAUGCCAUUCUCUUUUGCUUUCUAUUAUAUCGUAUUUUUUUCC------ ..(((((((---..((((..((((((......))))))..............((...........))..........))))....)))))))------ ( -11.60) >consensus ___GGACAA____UCGAUGCUGGUUGGCCCCACAAUCACUUUUCAAUUAAAUGGCAUUCUUUCCUCUUUGCCAUUAUUUUGUGUUUUUU_________ ....................((((((......))))))...........(((((((............)))))))....................... (-11.07 = -11.27 + 0.19)



| Location | 18,806,480 – 18,806,572 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 79.53 |
| Mean single sequence MFE | -16.97 |
| Consensus MFE | -10.49 |
| Energy contribution | -10.72 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.786521 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18806480 92 - 27905053 AAAGCAAAAAAAAAACACAAAAUAAUGGCAAAGAGGAAAGAAUGCCAUUUAAUUGAAAAGUGAUUGUGGGGCCAACCAGCAUCGA-G--UUGUCC--- ...((..........(((((...(((((((............)))))))..(((....)))..)))))((.....)).)).....-.--......--- ( -15.60) >DroSec_CAF1 31871 84 - 1 ---------AAAAAACACAAAAUAAUGGCAAAGAGGAAAGAAUGCCAUUUAAUUGAAAAGUGAUUGUGGGGCCAACCAGCAUCGGAG--UUGUCC--- ---------......(((((...(((((((............)))))))..(((....)))..)))))((((...((......))..--..))))--- ( -15.20) >DroSim_CAF1 34406 78 - 1 ---------AAAAAACACAAAAUAAUGGCAAAGAGGAAAGAAUGCCAUUUAAUUGAAAAGUGAUUGUGGGGCCAACCAGCAUCGA--------CA--- ---------........(((...(((((((............)))))))...)))....(((((.((.((.....)).))))).)--------).--- ( -14.10) >DroEre_CAF1 32407 84 - 1 AGAGC----AAAAAACACAAAAUAAUGGCAAAGAGGAAAGAAUGCCAUUUAAUUGAAAAGUGAUUGUGGGGCCAACCAG----------UUGCCCCCA ...((----((...((.(((...(((((((............)))))))...)))....))..))))((((.(((....----------))).)))). ( -19.20) >DroYak_CAF1 34170 94 - 1 GGAGC----AAAAAACACAAAAUAAUGGCAAAGAGGAAAGAAUGCCAUUUAAUUGAAAAGUGAUUGUGGGGCCAACCAACAUCGAGCUUCUGCCCCCU (((((----........(((...(((((((............)))))))...))).....((((..(((......)))..)))).)))))........ ( -20.50) >DroAna_CAF1 33334 89 - 1 ------GGAAAAAAAUACGAUAUAAUAGAAAGCAAAAGAGAAUGGCAUUUAAUUGAAAAGUGAUUGUGGGGCCAACCAGAAUCGAG---UUUUUCCGA ------(((((((.........((((.(((.((...........)).))).)))).....(((((.(((......))).)))))..---))))))).. ( -17.20) >consensus _________AAAAAACACAAAAUAAUGGCAAAGAGGAAAGAAUGCCAUUUAAUUGAAAAGUGAUUGUGGGGCCAACCAGCAUCGA____UUGUCC___ ...............(((((...(((((((............)))))))..(((....)))..)))))((.....))..................... (-10.49 = -10.72 + 0.22)


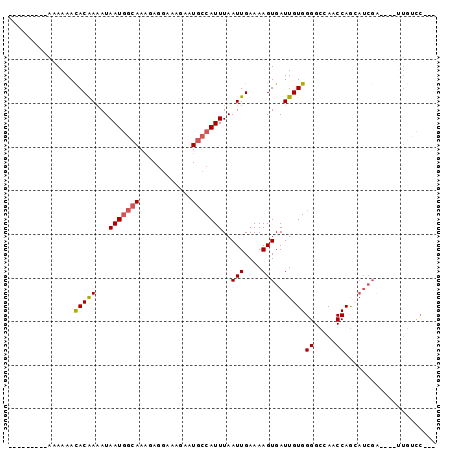
| Location | 18,806,493 – 18,806,583 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 83.05 |
| Mean single sequence MFE | -14.28 |
| Consensus MFE | -11.07 |
| Energy contribution | -11.27 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.870576 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18806493 90 + 27905053 CUGGUUGGCCCCACAAUCACUUUUCAAUUAAAUGGCAUUCUUUCCUCUUUGCCAUUAUUUUGUGUUUUUUUUUUGCUUUAGCCAUGGCUC .((((((......)))))).....(((...(((((((............)))))))...))).................(((....))). ( -16.00) >DroSec_CAF1 31885 74 + 1 CUGGUUGGCCCCACAAUCACUUUUCAAUUAAAUGGCAUUCUUUCCUCUUUGCCAUUAUUUUGUGUUUUUU----------------GCUC .((((((......)))))).....(((...(((((((............)))))))...)))........----------------.... ( -13.40) >DroSim_CAF1 34414 74 + 1 CUGGUUGGCCCCACAAUCACUUUUCAAUUAAAUGGCAUUCUUUCCUCUUUGCCAUUAUUUUGUGUUUUUU----------------GCUC .((((((......)))))).....(((...(((((((............)))))))...)))........----------------.... ( -13.40) >DroEre_CAF1 32416 86 + 1 CUGGUUGGCCCCACAAUCACUUUUCAAUUAAAUGGCAUUCUUUCCUCUUUGCCAUUAUUUUGUGUUUUUU----GCUCUAUGCAGCGCUC .((((((......))))))...........(((((((............))))))).....((((....(----((.....))))))).. ( -17.80) >DroYak_CAF1 34189 86 + 1 UUGGUUGGCCCCACAAUCACUUUUCAAUUAAAUGGCAUUCUUUCCUCUUUGCCAUUAUUUUGUGUUUUUU----GCUCCAUGCAACGCUC .((((((......))))))...........(((((((............))))))).....((((....(----((.....))))))).. ( -17.60) >DroAna_CAF1 33350 83 + 1 CUGGUUGGCCCCACAAUCACUUUUCAAUUAAAUGCCAUUCUCUUUUGCUUUCUAUUAUAUCGUAUUUUUUUCC-------UGCCUGGAUC .((((((......))))))...................................................(((-------.....))).. ( -7.50) >consensus CUGGUUGGCCCCACAAUCACUUUUCAAUUAAAUGGCAUUCUUUCCUCUUUGCCAUUAUUUUGUGUUUUUU__________UGCA__GCUC .((((((......))))))...........(((((((............))))))).................................. (-11.07 = -11.27 + 0.19)



| Location | 18,806,493 – 18,806,583 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 83.05 |
| Mean single sequence MFE | -14.12 |
| Consensus MFE | -10.49 |
| Energy contribution | -10.72 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.600765 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18806493 90 - 27905053 GAGCCAUGGCUAAAGCAAAAAAAAAACACAAAAUAAUGGCAAAGAGGAAAGAAUGCCAUUUAAUUGAAAAGUGAUUGUGGGGCCAACCAG ......(((((...((((................(((((((............)))))))..(((....)))..))))..)))))..... ( -17.60) >DroSec_CAF1 31885 74 - 1 GAGC----------------AAAAAACACAAAAUAAUGGCAAAGAGGAAAGAAUGCCAUUUAAUUGAAAAGUGAUUGUGGGGCCAACCAG ....----------------......(((((...(((((((............)))))))..(((....)))..)))))((.....)).. ( -13.00) >DroSim_CAF1 34414 74 - 1 GAGC----------------AAAAAACACAAAAUAAUGGCAAAGAGGAAAGAAUGCCAUUUAAUUGAAAAGUGAUUGUGGGGCCAACCAG ....----------------......(((((...(((((((............)))))))..(((....)))..)))))((.....)).. ( -13.00) >DroEre_CAF1 32416 86 - 1 GAGCGCUGCAUAGAGC----AAAAAACACAAAAUAAUGGCAAAGAGGAAAGAAUGCCAUUUAAUUGAAAAGUGAUUGUGGGGCCAACCAG ..(.(((.(((((..(----(.......(((...(((((((............)))))))...))).....)).))))).))))...... ( -16.20) >DroYak_CAF1 34189 86 - 1 GAGCGUUGCAUGGAGC----AAAAAACACAAAAUAAUGGCAAAGAGGAAAGAAUGCCAUUUAAUUGAAAAGUGAUUGUGGGGCCAACCAA .....((((.....))----))....(((((...(((((((............)))))))..(((....)))..)))))((.....)).. ( -15.80) >DroAna_CAF1 33350 83 - 1 GAUCCAGGCA-------GGAAAAAAAUACGAUAUAAUAGAAAGCAAAAGAGAAUGGCAUUUAAUUGAAAAGUGAUUGUGGGGCCAACCAG ..(((.....-------))).................................((((...(((((.......)))))....))))..... ( -9.10) >consensus GAGC__UGCA__________AAAAAACACAAAAUAAUGGCAAAGAGGAAAGAAUGCCAUUUAAUUGAAAAGUGAUUGUGGGGCCAACCAG ..........................(((((...(((((((............)))))))..(((....)))..)))))((.....)).. (-10.49 = -10.72 + 0.22)

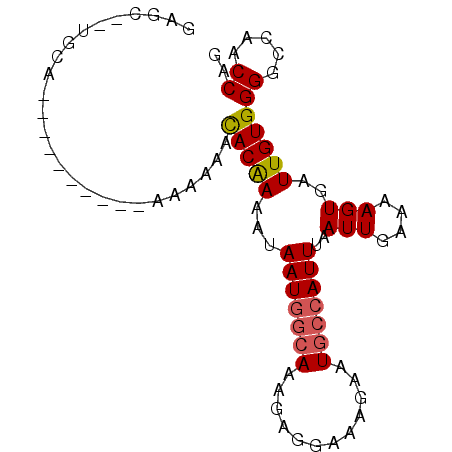

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:38:23 2006