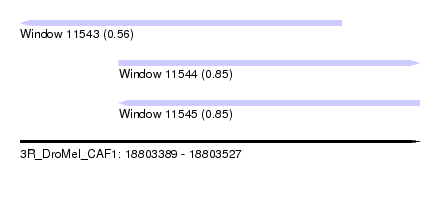
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,803,389 – 18,803,527 |
| Length | 138 |
| Max. P | 0.853377 |

| Location | 18,803,389 – 18,803,500 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 80.07 |
| Mean single sequence MFE | -50.92 |
| Consensus MFE | -32.97 |
| Energy contribution | -33.28 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.563353 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18803389 111 - 27905053 UUCCCGACAGCGACCACGAUGGUCGGGCAGAUAGCCGGGCUGGCAGGGGAAGUCGAUCAUGGCCACCAUCAUGGUCUUCUCGGUGGUGGUUUGGCCGGCGACCGGA--GGC---UC (((((..((((((((.....)))).(((.....)))..))))...))))).((((....(((((((((((((.(......).))))))))..))))).))))....--...---.. ( -50.10) >DroSec_CAF1 28784 111 - 1 UUCCCGACAACGACCACGAUGGUCGGGCAGAUAGCCCGGCUGGCAGGGGAAGUCGAUCAUGGCCACCAUCAUGGUCUUCUCGGUGGUGGUUUGGCCGGCGACCGUG--GGC---UC .............(((((..((((((((.....))))))))..........((((....(((((((((((((.(......).))))))))..))))).))))))))--)..---.. ( -48.80) >DroEre_CAF1 28014 105 - 1 UUCCCGACAGCGACCGCGAUGGUCGGGCAGAUAGCCCGGCUGGCAGGGGAAGUCGAUCAUGGCCACCAUCAUGGGCUUCUCGGUGGUGGC------GGCGCCCGGA--GAC---GC .(((((...(((.((((.((((((((((.....)))))))).((..((((((((.((.((((...)))).)).)))))))).)).)).))------))))).))).--)).---.. ( -49.30) >DroYak_CAF1 30919 111 - 1 UUCCCGACAGCGACCACGAUGGUCGGGCAGAUAGCCCGGCUGGCAGGGAAAGUCGAUCAUGGCCACCAUCAUGGGCUUCUCGGUGGUGGCAUUGCCGUCGCCCUGG--GAC---UC (((((..((((((((.....))))((((.....)))).))))...)))))((((.((.((((...)))).)).))))((((((.((((((......))))))))))--)).---.. ( -52.50) >DroMoj_CAF1 30774 108 - 1 CUCGCGGCACCGUCCGCGACUGUCGGGCAGGUAGCCCGGCUGGCAGGGGAAGUCGAUCAUGGCCACCUCCAUGAUCUUCUCGGUGGUGGCUGC-----UGAG-GAG--GGUCGCAC ...(((((.((.(((.(..(((((((((.....))))))).))..).)))....((((((((......))))))))(((((((..(...)..)-----))))-)))--)))))).. ( -52.80) >DroAna_CAF1 29881 114 - 1 UUCCCGGCACCGACCACGGUGGUCGGGCAGGUAGCCCGGCUGGCAGGGGAAGUCUAUCAUGGCCACCUCCAUGAUUUUCUCGGUAACGGGCUGUCCGCUGGCAGGCUUGAC--UGU ...(((((.((((((.....))))))...(((((((((....((..((((((...(((((((......))))))))))))).))..))))))).)))))))(((......)--)). ( -52.00) >consensus UUCCCGACAGCGACCACGAUGGUCGGGCAGAUAGCCCGGCUGGCAGGGGAAGUCGAUCAUGGCCACCAUCAUGGUCUUCUCGGUGGUGGCUUGGCCGGCGACCGGG__GAC___UC .........((.((((((.(((((((((.....))))))))).)..((((((...(((((((......))))))))))))).))))).)).......................... (-32.97 = -33.28 + 0.31)

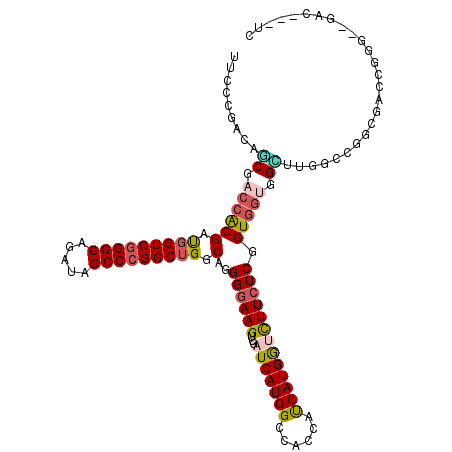

| Location | 18,803,423 – 18,803,527 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 79.91 |
| Mean single sequence MFE | -43.12 |
| Consensus MFE | -26.70 |
| Energy contribution | -26.70 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.851936 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18803423 104 + 27905053 AGACCAUGAUGGUGGCCAUGAUCGACUUCCCCUGCCAGCCCGGCUAUCUGCCCGACCAUCGUGGUCGCUGUCGGGAAAUCUGGUGAUUC--------GAUG-GCU-UCGUGACA .(((((((((((((((...((......))....))).....(((.....)))..))))))))))))((((((((...(((....)))))--------))))-)).-........ ( -39.90) >DroSec_CAF1 28818 107 + 1 AGACCAUGAUGGUGGCCAUGAUCGACUUCCCCUGCCAGCCGGGCUAUCUGCCCGACCAUCGUGGUCGUUGUCGGGAAAUCUGGUGAUCCGA-----CGAUG-GCU-UCGUGACA .(((((((((((((((...((......))....)))...(((((.....)))))))))))))))))((((((((...(((....)))))))-----)))).-...-........ ( -46.90) >DroEre_CAF1 28042 104 + 1 AGCCCAUGAUGGUGGCCAUGAUCGACUUCCCCUGCCAGCCGGGCUAUCUGCCCGACCAUCGCGGUCGCUGUCGGGAAAUCUGGUGAUUC--------GAUG-GCUUUCGUGAC- ....(((((....((((((((((.(((...((((.((((.((((.....))))((((.....)))))))).))))......))))))).--------.)))-))).)))))..- ( -41.80) >DroYak_CAF1 30953 106 + 1 AGCCCAUGAUGGUGGCCAUGAUCGACUUUCCCUGCCAGCCGGGCUAUCUGCCCGACCAUCGUGGUCGCUGUCGGGAAAUCUGGUGAUUC--------CGUGGGCUGCCGUGACA ((((((((..(((.((((.(((.....(((((.(.((((.((((.....))))((((.....)))))))).))))))))))))).))).--------))))))))......... ( -47.90) >DroMoj_CAF1 30805 90 + 1 AGAUCAUGGAGGUGGCCAUGAUCGACUUCCCCUGCCAGCCGGGCUACCUGCCCGACAGUCGCGGACGGUGCCGCGAGAUAUGGUAAACCC------------------------ .((((((((......)))))))).........((((((.(((((.....))))).)..((((((......))))))....))))).....------------------------ ( -38.20) >DroAna_CAF1 29918 112 + 1 AAAUCAUGGAGGUGGCCAUGAUAGACUUCCCCUGCCAGCCGGGCUACCUGCCCGACCACCGUGGUCGGUGCCGGGAAAUCUGGUAAACGGCAAAACCACUU-UCU-UCGUGAUA ..(((((((((((.(((....((((.(((((..((..(((((((.....))))((((.....))))))))).))))).))))......)))...)))....-..)-))))))). ( -44.00) >consensus AGACCAUGAUGGUGGCCAUGAUCGACUUCCCCUGCCAGCCGGGCUAUCUGCCCGACCAUCGUGGUCGCUGUCGGGAAAUCUGGUGAUCC________GAUG_GCU_UCGUGACA ...(((((((((((((...((......))....)))...(((((.....)))))))))))))))....((((((.....))))))............................. (-26.70 = -26.70 + 0.00)



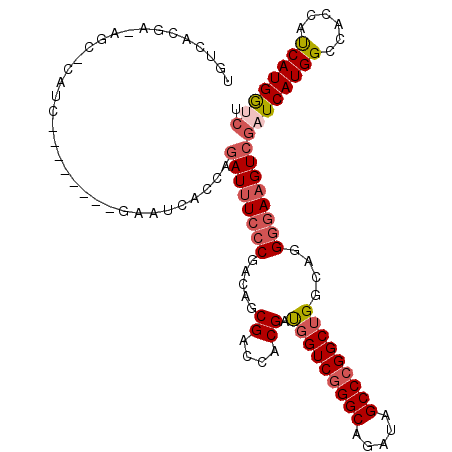
| Location | 18,803,423 – 18,803,527 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 79.91 |
| Mean single sequence MFE | -41.32 |
| Consensus MFE | -29.60 |
| Energy contribution | -30.52 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.853377 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18803423 104 - 27905053 UGUCACGA-AGC-CAUC--------GAAUCACCAGAUUUCCCGACAGCGACCACGAUGGUCGGGCAGAUAGCCGGGCUGGCAGGGGAAGUCGAUCAUGGCCACCAUCAUGGUCU .....(((-...-..))--------)........((((((((..((((((((.....)))).(((.....)))..))))....))))))))((((((((......)))))))). ( -38.80) >DroSec_CAF1 28818 107 - 1 UGUCACGA-AGC-CAUCG-----UCGGAUCACCAGAUUUCCCGACAACGACCACGAUGGUCGGGCAGAUAGCCCGGCUGGCAGGGGAAGUCGAUCAUGGCCACCAUCAUGGUCU ........-.((-(((.(-----((((....)).((((((((.....((....))..((((((((.....)))))))).....))))))))))).))))).(((.....))).. ( -40.40) >DroEre_CAF1 28042 104 - 1 -GUCACGAAAGC-CAUC--------GAAUCACCAGAUUUCCCGACAGCGACCGCGAUGGUCGGGCAGAUAGCCCGGCUGGCAGGGGAAGUCGAUCAUGGCCACCAUCAUGGGCU -........(((-(...--------.........((((((((..((((((((.....))))((((.....)))).))))....))))))))...(((((......))))))))) ( -40.90) >DroYak_CAF1 30953 106 - 1 UGUCACGGCAGCCCACG--------GAAUCACCAGAUUUCCCGACAGCGACCACGAUGGUCGGGCAGAUAGCCCGGCUGGCAGGGAAAGUCGAUCAUGGCCACCAUCAUGGGCU (((....)))(((((.(--------(.....)).((((((((..((((((((.....))))((((.....)))).))))...))).)))))....((((...))))..))))). ( -42.30) >DroMoj_CAF1 30805 90 - 1 ------------------------GGGUUUACCAUAUCUCGCGGCACCGUCCGCGACUGUCGGGCAGGUAGCCCGGCUGGCAGGGGAAGUCGAUCAUGGCCACCUCCAUGAUCU ------------------------((.((..((.....((((((......))))))(.(((((((.....))))))).)...))..)).))((((((((......)))))))). ( -39.90) >DroAna_CAF1 29918 112 - 1 UAUCACGA-AGA-AAGUGGUUUUGCCGUUUACCAGAUUUCCCGGCACCGACCACGGUGGUCGGGCAGGUAGCCCGGCUGGCAGGGGAAGUCUAUCAUGGCCACCUCCAUGAUUU .(((((..-...-..))))).............(((((((((.((((((....))))((((((((.....)))))))).))..)))))))))(((((((......))))))).. ( -45.60) >consensus UGUCACGA_AGC_CAUC________GAAUCACCAGAUUUCCCGACAGCGACCACGAUGGUCGGGCAGAUAGCCCGGCUGGCAGGGGAAGUCGAUCAUGGCCACCAUCAUGGUCU ..................................((((((((.....((....)).(((((((((.....)))))))))....))))))))((((((((......)))))))). (-29.60 = -30.52 + 0.92)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:38:18 2006