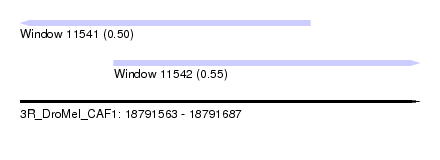
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,791,563 – 18,791,687 |
| Length | 124 |
| Max. P | 0.549731 |

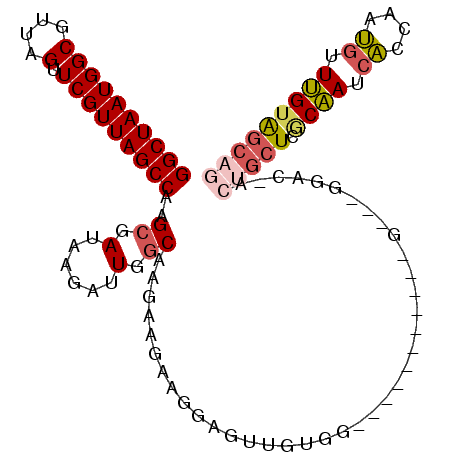
| Location | 18,791,563 – 18,791,653 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 82.22 |
| Mean single sequence MFE | -21.22 |
| Consensus MFE | -14.17 |
| Energy contribution | -14.70 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.67 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18791563 90 - 27905053 CCCCCCUACAACUCCUUCUUGCCAAUCUUAUCGCUUGGCUAACGAACUAACGCCAUUAGCCAACGUGGCCGGUGCGACAACCAGUG----GCUA ....................((((......((((((((((((((......))...)))))))).))))..(((......)))..))----)).. ( -21.80) >DroSec_CAF1 16848 90 - 1 CCCCGCAACUCCUUCUCCUUGCCAAUCUUAUCGCUUGGCUAACGAACUAACGCCAUUAGCCAACGCGGCCGGUGCGACAACCAGUG----GCUA ....................((((......((((((((((((((......))...)))))))).))))..(((......)))..))----)).. ( -23.70) >DroSim_CAF1 19211 90 - 1 CCCCACAACUCCUUCUUCUUGCCAAUCUUAUCGCUUGGCUAACGAACUAACGCCAUUAGCCAACGCGGCCGGUGCGACAACCAGUG----GCUA ....................((((......((((((((((((((......))...)))))))).))))..(((......)))..))----)).. ( -23.70) >DroEre_CAF1 16416 87 - 1 CCGCAAAACUCCU---UCUCGCCAAUCUUAUCGCUUGGCUAACGAACUAACGCCAUUAGCCAACGCGGCCGGUGCGACAACCAGUG----GCUA .............---....((((......((((((((((((((......))...)))))))).))))..(((......)))..))----)).. ( -24.00) >DroWil_CAF1 17158 68 - 1 -------------------UGGCAAUCUUAUCGCUUGGCUAACGAAGUAACGCCAUUAGGCAACGCGU---UUGCGACAACAAACA----ACAA -------------------..((.........((((.(....).))))...(((....)))...))((---(((......))))).----.... ( -13.40) >DroYak_CAF1 18576 91 - 1 CCGCACAACUCCU---UCUUGCCAAUCUUAUCGCUUGGCUAACGAACUAACGCCAUUAGCCAACGCGGCCGGUGCGACAACCAGUGACCAGCUA ...(((.......---..............((((((((((((((......))...)))))))).))))..(((......))).)))........ ( -20.70) >consensus CCCCACAACUCCU___UCUUGCCAAUCUUAUCGCUUGGCUAACGAACUAACGCCAUUAGCCAACGCGGCCGGUGCGACAACCAGUG____GCUA ....................((.........(((((((((((((......))...)))))))).)))...(((......)))........)).. (-14.17 = -14.70 + 0.53)

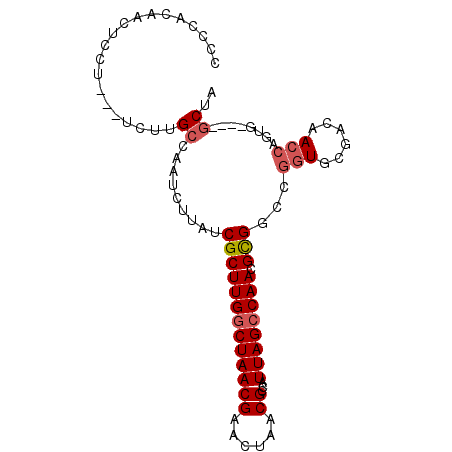
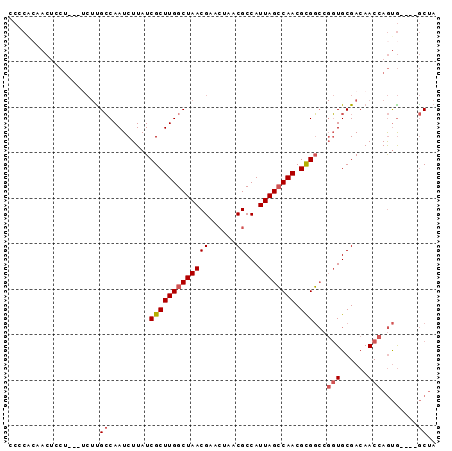
| Location | 18,791,592 – 18,791,687 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 79.33 |
| Mean single sequence MFE | -30.72 |
| Consensus MFE | -17.54 |
| Energy contribution | -18.10 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.549731 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18791592 95 + 27905053 GGCUAAUGGCGUUAGUUCGUUAGCCAAGCGAUAAGAUUGGCAAGAAGGAGUUGUAGGGG-----------G---GGAGUUGUGCUCGCAAUCACCAAUGUUUGUAGCAG ((((((((((....).)))))))))..((.(((((((((((((.......)))..((..-----------(---.(((.....))).)..)).))))).))))).)).. ( -26.10) >DroPse_CAF1 23980 109 + 1 GGCUAAUGGCGUUAGUUCGUUAGCCAAGCGAUAAGAUUGCCAGCCGAAAGGGGGUCCGGUUCAAAGAACAGGGAGGGCAAGGCAUCCCCAUCUCCCAAUCUGGCGGCAG ((((((((((....).)))))))))............((((.(((((..(((((....((((...)))).((((..((...)).))))...)))))..)).))))))). ( -40.20) >DroSec_CAF1 16877 95 + 1 GGCUAAUGGCGUUAGUUCGUUAGCCAAGCGAUAAGAUUGGCAAGGAGAAGGAGUUGCGG-----------G---GGACGACUGCUCGCAAUCACCAAUGUUUGUAGCAG .((((..((((((..(((.((.(((((.(.....).))))))).)))..((.(((((((-----------(---.(.....).))))))))..))))))))..)))).. ( -29.50) >DroSim_CAF1 19240 95 + 1 GGCUAAUGGCGUUAGUUCGUUAGCCAAGCGAUAAGAUUGGCAAGAAGAAGGAGUUGUGG-----------G---GGACGACUGCUCGCAAUCACCAAUGUUUGUAGCAG ((((((((((....).)))))))))..((.((((((((((...((.....((((.((.(-----------.---...).)).))))....)).))))).))))).)).. ( -30.30) >DroEre_CAF1 16445 89 + 1 GGCUAAUGGCGUUAGUUCGUUAGCCAAGCGAUAAGAUUGGCGAGA---AGGAGUUUUGC-----------G---GG---ACUGCUCACAAUCACCAAUGUUUGUGGCAG ((((((((((....).)))))))))..((.((((((((((.((..---..((((.....-----------.---..---...))))....)).))))).))))).)).. ( -30.60) >DroYak_CAF1 18609 90 + 1 GGCUAAUGGCGUUAGUUCGUUAGCCAAGCGAUAAGAUUGGCAAGA---AGGAGUUGUGC-----------G---GGA--ACCGCUCACAAUCACCAAUGUUUGUAGCAG ((((((((((....).)))))))))..((.((((((((((.....---..((((.((..-----------.---...--)).)))).......))))).))))).)).. ( -27.64) >consensus GGCUAAUGGCGUUAGUUCGUUAGCCAAGCGAUAAGAUUGGCAAGAAGAAGGAGUUGUGG___________G___GGAC_ACUGCUCGCAAUCACCAAUGUUUGUAGCAG ((((((((((....).)))))))))..((.(......).)).......................................(((((.((((.((....)).))))))))) (-17.54 = -18.10 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:38:15 2006