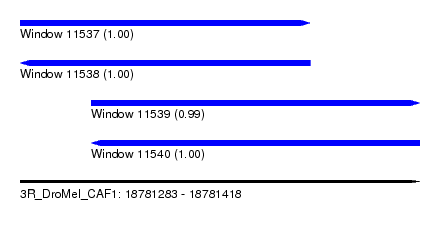
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,781,283 – 18,781,418 |
| Length | 135 |
| Max. P | 0.999766 |

| Location | 18,781,283 – 18,781,381 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 90.95 |
| Mean single sequence MFE | -30.90 |
| Consensus MFE | -27.37 |
| Energy contribution | -27.70 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.58 |
| Structure conservation index | 0.89 |
| SVM decision value | 3.39 |
| SVM RNA-class probability | 0.999136 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18781283 98 + 27905053 C---------CCUUACUUUUUCGCUUGCCAACAGCAUUUGUCUUUUGACAUUUGCUACUUGCACUCUCGCCGACUUCAUU-UUAGCCGGGGCCAAAAGAUGUCGGCGA .---------...............(((....((((..((((....))))..))))....)))...((((((((.((.((-((.((....)).)))))).)))))))) ( -32.80) >DroSec_CAF1 6661 98 + 1 C---------CCUUACUUUUUCGCUUGCCAACAGCAUUUGUCUUUUGACAUUUGCUACUUGCACUCUCGCCGACUUCAUU-UUAGCCGGGGCCAAAAGAUGUCGGCGA .---------...............(((....((((..((((....))))..))))....)))...((((((((.((.((-((.((....)).)))))).)))))))) ( -32.80) >DroSim_CAF1 8370 98 + 1 C---------CCUUACUUUUUCGCUUGCCAACAGCAUUUCUCUUUUGACAUUUGCUACUUGCACUCUCGCCGACUUCAUU-UUAGCCGGGGCCAAAAGAUGUCGGCGA .---------...............(((....((((..(.((....)).)..))))....)))...((((((((.((.((-((.((....)).)))))).)))))))) ( -26.80) >DroEre_CAF1 6534 101 + 1 C-----GUUUCCGCACUUUUUCGCUUGCCAACAGCAUUUGUCUUUUGACAUUUGCUACUUGCACUCUCGCCGACUUCAUU-UUAGCCGG-GCCAAAAGACGUCGGCGA .-----......((........)).(((....((((..((((....))))..))))....)))...((((((((.((.((-((.((...-)).)))))).)))))))) ( -30.60) >DroYak_CAF1 7152 107 + 1 CAUUUGGUUUACGUAGUUUUUCGCUUGCCAACAGCAUUUGUCUUUUGACAUUUGCUACUUGCACUCUCGCCGACUUCAUU-UUAGCCGGGGCCAAAAGAUGUCGGCGA ...(((((...((........))...))))).((((..((((....))))..))))..........((((((((.((.((-((.((....)).)))))).)))))))) ( -35.90) >DroAna_CAF1 7760 95 + 1 C----------CGUACUUUUUCGCUUGCCAACAGCAUUUGUCUUUUGACAUUUGCUACUUGCACUCUCGCCGACUUCAUUUUUAGCAAGAGC---AAGAUGUCGGCGC .----------..............(((....((((..((((....))))..))))....)))....(((((((.....((((.((....))---)))).))))))). ( -26.50) >consensus C_________CCGUACUUUUUCGCUUGCCAACAGCAUUUGUCUUUUGACAUUUGCUACUUGCACUCUCGCCGACUUCAUU_UUAGCCGGGGCCAAAAGAUGUCGGCGA .........................(((....((((..((((....))))..))))....)))...((((((((.((.((....((....))...)))).)))))))) (-27.37 = -27.70 + 0.33)

| Location | 18,781,283 – 18,781,381 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 90.95 |
| Mean single sequence MFE | -37.43 |
| Consensus MFE | -33.85 |
| Energy contribution | -34.68 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.00 |
| Mean z-score | -4.77 |
| Structure conservation index | 0.90 |
| SVM decision value | 4.03 |
| SVM RNA-class probability | 0.999766 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18781283 98 - 27905053 UCGCCGACAUCUUUUGGCCCCGGCUAA-AAUGAAGUCGGCGAGAGUGCAAGUAGCAAAUGUCAAAAGACAAAUGCUGUUGGCAAGCGAAAAAGUAAGG---------G ((((((((.(((((((((....)))))-)).)).))))))))(..(((...(((((..((((....))))..)))))...)))..)............---------. ( -39.60) >DroSec_CAF1 6661 98 - 1 UCGCCGACAUCUUUUGGCCCCGGCUAA-AAUGAAGUCGGCGAGAGUGCAAGUAGCAAAUGUCAAAAGACAAAUGCUGUUGGCAAGCGAAAAAGUAAGG---------G ((((((((.(((((((((....)))))-)).)).))))))))(..(((...(((((..((((....))))..)))))...)))..)............---------. ( -39.60) >DroSim_CAF1 8370 98 - 1 UCGCCGACAUCUUUUGGCCCCGGCUAA-AAUGAAGUCGGCGAGAGUGCAAGUAGCAAAUGUCAAAAGAGAAAUGCUGUUGGCAAGCGAAAAAGUAAGG---------G ((((((((.(((((((((....)))))-)).)).))))))))(..(((...(((((..(.((....)).)..)))))...)))..)............---------. ( -35.70) >DroEre_CAF1 6534 101 - 1 UCGCCGACGUCUUUUGGC-CCGGCUAA-AAUGAAGUCGGCGAGAGUGCAAGUAGCAAAUGUCAAAAGACAAAUGCUGUUGGCAAGCGAAAAAGUGCGGAAAC-----G ((((((((.(((((((((-...)))))-)).)).))))))))(..(((...(((((..((((....))))..)))))...)))..).........((....)-----) ( -39.00) >DroYak_CAF1 7152 107 - 1 UCGCCGACAUCUUUUGGCCCCGGCUAA-AAUGAAGUCGGCGAGAGUGCAAGUAGCAAAUGUCAAAAGACAAAUGCUGUUGGCAAGCGAAAAACUACGUAAACCAAAUG ((((((((.(((((((((....)))))-)).)).))))))))(..(((...(((((..((((....))))..)))))...)))..)...................... ( -39.60) >DroAna_CAF1 7760 95 - 1 GCGCCGACAUCUU---GCUCUUGCUAAAAAUGAAGUCGGCGAGAGUGCAAGUAGCAAAUGUCAAAAGACAAAUGCUGUUGGCAAGCGAAAAAGUACG----------G ((((((((..(((---((((((((..(........)..))))))..))))).((((..((((....))))..))))))))))..))...........----------. ( -31.10) >consensus UCGCCGACAUCUUUUGGCCCCGGCUAA_AAUGAAGUCGGCGAGAGUGCAAGUAGCAAAUGUCAAAAGACAAAUGCUGUUGGCAAGCGAAAAAGUAAGG_________G ((((((((.((..(((((....)))))....)).))))))))(..(((...(((((..((((....))))..)))))...)))..)...................... (-33.85 = -34.68 + 0.83)

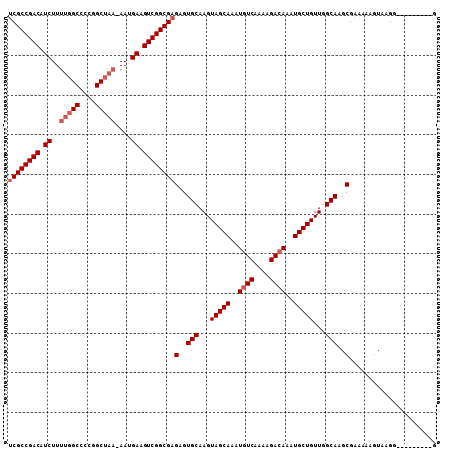
| Location | 18,781,307 – 18,781,418 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 95.28 |
| Mean single sequence MFE | -32.05 |
| Consensus MFE | -27.47 |
| Energy contribution | -27.52 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.19 |
| SVM RNA-class probability | 0.989891 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18781307 111 + 27905053 GCAUUUGUCUUUUGACAUUUGCUACUUGCACUCUCGCCGACUUCAUU-UUAGCCGGGGCCAAAAGAUGUCGGCGAUUGAUUCAAUUUGUUGCC-ACUCUGUUUACAAAGCGUU (((..((((....))))..))).....((.(..((((((((.((.((-((.((....)).)))))).))))))))..)......(((((.((.-.....))..)))))))... ( -34.60) >DroSec_CAF1 6685 111 + 1 GCAUUUGUCUUUUGACAUUUGCUACUUGCACUCUCGCCGACUUCAUU-UUAGCCGGGGCCAAAAGAUGUCGGCGAUUGAUUCAAUUUGUUGCC-ACUCUGUUUACAAAGCGUU (((..((((....))))..))).....((.(..((((((((.((.((-((.((....)).)))))).))))))))..)......(((((.((.-.....))..)))))))... ( -34.60) >DroSim_CAF1 8394 111 + 1 GCAUUUCUCUUUUGACAUUUGCUACUUGCACUCUCGCCGACUUCAUU-UUAGCCGGGGCCAAAAGAUGUCGGCGAUUGAUUCAAUUUGUUGCC-ACUCUGUUUACAAAGCGUU ((.........((((....(((.....)))(..((((((((.((.((-((.((....)).)))))).))))))))..)..))))(((((.((.-.....))..)))))))... ( -28.80) >DroEre_CAF1 6562 110 + 1 GCAUUUGUCUUUUGACAUUUGCUACUUGCACUCUCGCCGACUUCAUU-UUAGCCGG-GCCAAAAGACGUCGGCGAUUGAUUCAAUUUGUUGCC-ACUCUGUUUACAAAGCGUU (((..((((....))))..))).....((.(..((((((((.((.((-((.((...-)).)))))).))))))))..)......(((((.((.-.....))..)))))))... ( -31.60) >DroYak_CAF1 7185 111 + 1 GCAUUUGUCUUUUGACAUUUGCUACUUGCACUCUCGCCGACUUCAUU-UUAGCCGGGGCCAAAAGAUGUCGGCGAUUGAUUCAAUUUGUUGCC-ACUCUGUUUACAAAGCGUU (((..((((....))))..))).....((.(..((((((((.((.((-((.((....)).)))))).))))))))..)......(((((.((.-.....))..)))))))... ( -34.60) >DroAna_CAF1 7783 110 + 1 GCAUUUGUCUUUUGACAUUUGCUACUUGCACUCUCGCCGACUUCAUUUUUAGCAAGAGC---AAGAUGUCGGCGCUUGAUUCAAUUUGUUCCAAACUCUGUUUGCAAAGUGUU (((..((((....))))..))).....((((((.(((((((.....((((.((....))---)))).)))))))...)).....(((((..((.....))...))))))))). ( -28.10) >consensus GCAUUUGUCUUUUGACAUUUGCUACUUGCACUCUCGCCGACUUCAUU_UUAGCCGGGGCCAAAAGAUGUCGGCGAUUGAUUCAAUUUGUUGCC_ACUCUGUUUACAAAGCGUU (((..((((....))))..))).....((.(..((((((((.((.((....((....))...)))).))))))))..)......(((((..............)))))))... (-27.47 = -27.52 + 0.06)
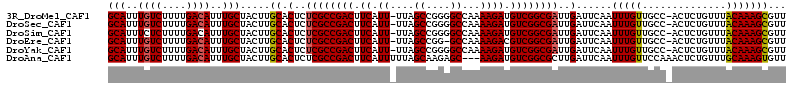
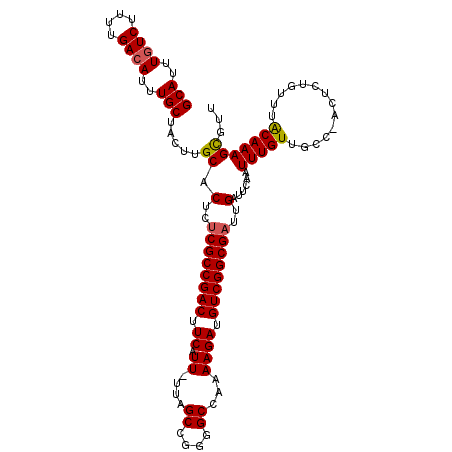
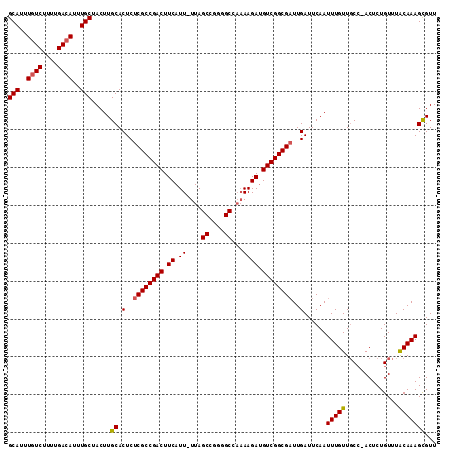
| Location | 18,781,307 – 18,781,418 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 95.28 |
| Mean single sequence MFE | -37.40 |
| Consensus MFE | -31.12 |
| Energy contribution | -31.82 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.03 |
| Mean z-score | -4.45 |
| Structure conservation index | 0.83 |
| SVM decision value | 3.29 |
| SVM RNA-class probability | 0.998931 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18781307 111 - 27905053 AACGCUUUGUAAACAGAGU-GGCAACAAAUUGAAUCAAUCGCCGACAUCUUUUGGCCCCGGCUAA-AAUGAAGUCGGCGAGAGUGCAAGUAGCAAAUGUCAAAAGACAAAUGC ..(((((((....))))))-)........(((...(..((((((((.(((((((((....)))))-)).)).))))))))..)..)))...(((..((((....))))..))) ( -40.50) >DroSec_CAF1 6685 111 - 1 AACGCUUUGUAAACAGAGU-GGCAACAAAUUGAAUCAAUCGCCGACAUCUUUUGGCCCCGGCUAA-AAUGAAGUCGGCGAGAGUGCAAGUAGCAAAUGUCAAAAGACAAAUGC ..(((((((....))))))-)........(((...(..((((((((.(((((((((....)))))-)).)).))))))))..)..)))...(((..((((....))))..))) ( -40.50) >DroSim_CAF1 8394 111 - 1 AACGCUUUGUAAACAGAGU-GGCAACAAAUUGAAUCAAUCGCCGACAUCUUUUGGCCCCGGCUAA-AAUGAAGUCGGCGAGAGUGCAAGUAGCAAAUGUCAAAAGAGAAAUGC ..(((((((....))))))-)........((((.....((((((((.(((((((((....)))))-)).)).))))))))...(((.....)))....))))........... ( -37.10) >DroEre_CAF1 6562 110 - 1 AACGCUUUGUAAACAGAGU-GGCAACAAAUUGAAUCAAUCGCCGACGUCUUUUGGC-CCGGCUAA-AAUGAAGUCGGCGAGAGUGCAAGUAGCAAAUGUCAAAAGACAAAUGC ..(((((((....))))))-)........(((...(..((((((((.(((((((((-...)))))-)).)).))))))))..)..)))...(((..((((....))))..))) ( -37.50) >DroYak_CAF1 7185 111 - 1 AACGCUUUGUAAACAGAGU-GGCAACAAAUUGAAUCAAUCGCCGACAUCUUUUGGCCCCGGCUAA-AAUGAAGUCGGCGAGAGUGCAAGUAGCAAAUGUCAAAAGACAAAUGC ..(((((((....))))))-)........(((...(..((((((((.(((((((((....)))))-)).)).))))))))..)..)))...(((..((((....))))..))) ( -40.50) >DroAna_CAF1 7783 110 - 1 AACACUUUGCAAACAGAGUUUGGAACAAAUUGAAUCAAGCGCCGACAUCUU---GCUCUUGCUAAAAAUGAAGUCGGCGAGAGUGCAAGUAGCAAAUGUCAAAAGACAAAUGC ...((((.(((..(...((((((..(.....)..))))))((((((.((..---((....)).......)).))))))..)..))))))).(((..((((....))))..))) ( -28.30) >consensus AACGCUUUGUAAACAGAGU_GGCAACAAAUUGAAUCAAUCGCCGACAUCUUUUGGCCCCGGCUAA_AAUGAAGUCGGCGAGAGUGCAAGUAGCAAAUGUCAAAAGACAAAUGC ...((((((....))))))..........(((......((((((((.((..(((((....)))))....)).)))))))).....)))...(((..((((....))))..))) (-31.12 = -31.82 + 0.70)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:38:14 2006