| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,764,854 – 18,764,954 |
| Length | 100 |
| Max. P | 0.833511 |

| Location | 18,764,854 – 18,764,954 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 81.37 |
| Mean single sequence MFE | -24.86 |
| Consensus MFE | -17.58 |
| Energy contribution | -17.34 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.583932 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18764854 100 + 27905053 AUGGUGCAAAUGGGGA-------AAAUGGGGAAAAUGGAGAAAAUUGGCAUGGGGCUUUGGUUUUUCUAAGCCGCAUUCCCGUUGUGAUUAAAUGCACGUAAUUAUA ...(((((((((((((-------...((.((....((((((((...(((.....)))....))))))))..)).)))))))))).........)))))......... ( -27.30) >DroSec_CAF1 23249 92 + 1 AUGGCGCAAAUGGGGA-------AAAUGGGGAG--------AAAUUGGCAUGGGGCUUUGGUUUUUCUAAGCCGCAUUCCCGUUGUGAUUAAAUGCACGUAAUUAUA ....(((((.((((((-------..(((..((.--------...))..)))(.(((((.((....)).))))).).))))))))))).................... ( -23.00) >DroSim_CAF1 23882 92 + 1 AUGGCGCAAAUGGGGA-------AAAUGGGGAA--------AAAUUGGCAUGGGGCUUUGGUUUUUCUAAGCCGCAUUCCCGUUGUGAUUAAAUGCACGUAAUUAUA ....(((((...((((-------(..(((((((--------((((((((.....)))..)))))))))...)))..))))).))))).................... ( -21.80) >DroEre_CAF1 23453 82 + 1 AU-------------------------GGUGAAAAUGGGGAAAAUGGGCAUGGGGCUUUGGUUUUUCUAAGCCGCAUUCCCGUUGUGAUUAAAUGCACGUAAUUACA ..-------------------------...............((((((.(((.(((((.((....)).))))).))).))))))(((((((........))))))). ( -24.40) >DroYak_CAF1 18107 107 + 1 AUGCUACAAAUGGUAAAAUGGUGAAAUGGUGAAAAUGAGGAAAAUGGGCAUGGGGCUUUGGUUUUUCUAAGCCGCAUUCCCGUUGUGAUUAAAUGCACGUAAUUACA .(((((....))))).....((((....(((....(((....((((((.(((.(((((.((....)).))))).))).))))))....)))....)))....)))). ( -27.80) >consensus AUGGCGCAAAUGGGGA_______AAAUGGGGAAAAUG__GAAAAUUGGCAUGGGGCUUUGGUUUUUCUAAGCCGCAUUCCCGUUGUGAUUAAAUGCACGUAAUUAUA ..........................................(((.((.(((.(((((.((....)).))))).))).)).)))(((((((........))))))). (-17.58 = -17.34 + -0.24)



| Location | 18,764,854 – 18,764,954 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 81.37 |
| Mean single sequence MFE | -17.05 |
| Consensus MFE | -12.14 |
| Energy contribution | -12.14 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.833511 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18764854 100 - 27905053 UAUAAUUACGUGCAUUUAAUCACAACGGGAAUGCGGCUUAGAAAAACCAAAGCCCCAUGCCAAUUUUCUCCAUUUUCCCCAUUU-------UCCCCAUUUGCACCAU .........(((((............(((((((.(((((.(......).))))).))).................)))).....-------........)))))... ( -16.83) >DroSec_CAF1 23249 92 - 1 UAUAAUUACGUGCAUUUAAUCACAACGGGAAUGCGGCUUAGAAAAACCAAAGCCCCAUGCCAAUUU--------CUCCCCAUUU-------UCCCCAUUUGCGCCAU .........(((((............(((((((.(((((.(......).))))).)))........--------.)))).....-------........)))))... ( -17.11) >DroSim_CAF1 23882 92 - 1 UAUAAUUACGUGCAUUUAAUCACAACGGGAAUGCGGCUUAGAAAAACCAAAGCCCCAUGCCAAUUU--------UUCCCCAUUU-------UCCCCAUUUGCGCCAU .........(((((............(((((((.(((((.(......).))))).)))........--------.)))).....-------........)))))... ( -17.11) >DroEre_CAF1 23453 82 - 1 UGUAAUUACGUGCAUUUAAUCACAACGGGAAUGCGGCUUAGAAAAACCAAAGCCCCAUGCCCAUUUUCCCCAUUUUCACC-------------------------AU .........(((........)))...(((.(((.(((((.(......).))))).))).)))..................-------------------------.. ( -16.10) >DroYak_CAF1 18107 107 - 1 UGUAAUUACGUGCAUUUAAUCACAACGGGAAUGCGGCUUAGAAAAACCAAAGCCCCAUGCCCAUUUUCCUCAUUUUCACCAUUUCACCAUUUUACCAUUUGUAGCAU .........((((.............(((.(((.(((((.(......).))))).))).)))..............................(((.....))))))) ( -18.10) >consensus UAUAAUUACGUGCAUUUAAUCACAACGGGAAUGCGGCUUAGAAAAACCAAAGCCCCAUGCCAAUUUUC__CAUUUUCCCCAUUU_______UCCCCAUUUGCACCAU .........(((........)))....((.(((.(((((.(......).))))).))).)).............................................. (-12.14 = -12.14 + -0.00)

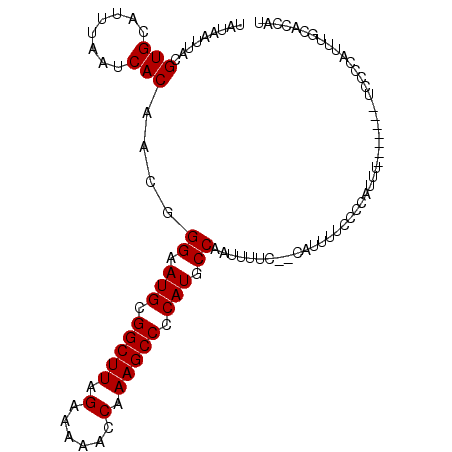

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:37:55 2006