
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,748,440 – 18,748,620 |
| Length | 180 |
| Max. P | 0.999437 |

| Location | 18,748,440 – 18,748,553 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 98.24 |
| Mean single sequence MFE | -33.28 |
| Consensus MFE | -32.58 |
| Energy contribution | -32.78 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.98 |
| SVM decision value | 3.60 |
| SVM RNA-class probability | 0.999437 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18748440 113 + 27905053 UAUAUUGGCAGUUAUGCUGCAAAUUUAUUGGCAUGCUUUGUAAACUGGGUUAGUUGCAUAU--AUCUCGCAGCCGCAAGCUGCGAGCCAAACGAUUAACGUUGAGUAACCCAAAU .......((((.....))))...(((((.((....))..))))).(((((((.((......--..(((((((.(....))))))))...((((.....)))))).)))))))... ( -33.60) >DroSec_CAF1 7188 113 + 1 UAUAUUGGCAGUUAUGCUGCAAAUUUAUUGGCAUGCUUUGUAAACUGGGUUAGUUGCAUAU--AUCUCGCAGCCGCAAGCUGCGAGCCAAACGAUUAACGUUGAGUAACCCAAAU .......((((.....))))...(((((.((....))..))))).(((((((.((......--..(((((((.(....))))))))...((((.....)))))).)))))))... ( -33.60) >DroEre_CAF1 7250 113 + 1 UAUAUUGGCAGUUAUGCUGAAAAUUUAUUGGCAUGCUUUGUAAACUGGGUUAGUUGCAUAU--AUCUCGCAGCCGCAAGCUGCGAGCCAAACGAUUAACGUUGAGUAACCCAAAU .......((((.(((((..(.......)..)))))..))))....(((((((.((......--..(((((((.(....))))))))...((((.....)))))).)))))))... ( -35.20) >DroYak_CAF1 1258 113 + 1 UAUAUUGGCAGUUAUGCUGCAAAUUUAUUGGCAUGCUUUGUAAACUGGGUUAGUUGCAUAU--AUCUCGCAGCCGCAAGCUGCGAGCCAAACGAUUAACGUUGAGUAACCCAAAU .......((((.....))))...(((((.((....))..))))).(((((((.((......--..(((((((.(....))))))))...((((.....)))))).)))))))... ( -33.60) >DroAna_CAF1 1423 115 + 1 UAUAUUGGCAGUUAUGCUGCAAAUUUAUUGGCAUGCUUUGUAAACUGGGUUAGUUGCAUAUACAUCUCGCAGCCGCAAGCUGCGCGGCAAACGAUUAACGUUGAGUAACCCAAAU .......((((.....))))...(((((.((....))..))))).(((((((.((.........((.(((((.(....)))))).))..((((.....)))))).)))))))... ( -30.40) >consensus UAUAUUGGCAGUUAUGCUGCAAAUUUAUUGGCAUGCUUUGUAAACUGGGUUAGUUGCAUAU__AUCUCGCAGCCGCAAGCUGCGAGCCAAACGAUUAACGUUGAGUAACCCAAAU .......((((.((((((...........))))))..))))....(((((((.((..........(((((((.(....))))))))...((((.....)))))).)))))))... (-32.58 = -32.78 + 0.20)



| Location | 18,748,440 – 18,748,553 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 98.24 |
| Mean single sequence MFE | -30.90 |
| Consensus MFE | -29.12 |
| Energy contribution | -29.72 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.94 |
| SVM decision value | 3.36 |
| SVM RNA-class probability | 0.999080 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18748440 113 - 27905053 AUUUGGGUUACUCAACGUUAAUCGUUUGGCUCGCAGCUUGCGGCUGCGAGAU--AUAUGCAACUAACCCAGUUUACAAAGCAUGCCAAUAAAUUUGCAGCAUAACUGCCAAUAUA ..((((((((((.((((.....)))).))((((((((.....))))))))..--.........))))))))....................(((.((((.....)))).)))... ( -32.10) >DroSec_CAF1 7188 113 - 1 AUUUGGGUUACUCAACGUUAAUCGUUUGGCUCGCAGCUUGCGGCUGCGAGAU--AUAUGCAACUAACCCAGUUUACAAAGCAUGCCAAUAAAUUUGCAGCAUAACUGCCAAUAUA ..((((((((((.((((.....)))).))((((((((.....))))))))..--.........))))))))....................(((.((((.....)))).)))... ( -32.10) >DroEre_CAF1 7250 113 - 1 AUUUGGGUUACUCAACGUUAAUCGUUUGGCUCGCAGCUUGCGGCUGCGAGAU--AUAUGCAACUAACCCAGUUUACAAAGCAUGCCAAUAAAUUUUCAGCAUAACUGCCAAUAUA ..((((((((((.((((.....)))).))((((((((.....))))))))..--.........))))))))........((((((.............)))....)))....... ( -29.22) >DroYak_CAF1 1258 113 - 1 AUUUGGGUUACUCAACGUUAAUCGUUUGGCUCGCAGCUUGCGGCUGCGAGAU--AUAUGCAACUAACCCAGUUUACAAAGCAUGCCAAUAAAUUUGCAGCAUAACUGCCAAUAUA ..((((((((((.((((.....)))).))((((((((.....))))))))..--.........))))))))....................(((.((((.....)))).)))... ( -32.10) >DroAna_CAF1 1423 115 - 1 AUUUGGGUUACUCAACGUUAAUCGUUUGCCGCGCAGCUUGCGGCUGCGAGAUGUAUAUGCAACUAACCCAGUUUACAAAGCAUGCCAAUAAAUUUGCAGCAUAACUGCCAAUAUA ..((((((((..(((((((..((((..((((((.....)))))).)))))))))...))....))))))))....................(((.((((.....)))).)))... ( -29.00) >consensus AUUUGGGUUACUCAACGUUAAUCGUUUGGCUCGCAGCUUGCGGCUGCGAGAU__AUAUGCAACUAACCCAGUUUACAAAGCAUGCCAAUAAAUUUGCAGCAUAACUGCCAAUAUA ..((((((((((.((((.....)))).))((((((((.....)))))))).............))))))))....................(((.((((.....)))).)))... (-29.12 = -29.72 + 0.60)



| Location | 18,748,475 – 18,748,587 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 90.54 |
| Mean single sequence MFE | -33.42 |
| Consensus MFE | -30.77 |
| Energy contribution | -30.77 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.45 |
| SVM RNA-class probability | 0.994117 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18748475 112 + 27905053 CUUUGUAAACUGGGUUAGUUGCAUAUAUCUCGCAGCCGCAAGCUGCGAGCCAAACGAUUAACGUUGAGUAACCCAAAUUGAUGUCACCACCCAUCGACCGCAGCUAACAGCC------ .............(((((((((......(((((((.(....))))))))...((((.....))))............((((((........))))))..)))))))))....------ ( -33.50) >DroSec_CAF1 7223 118 + 1 CUUUGUAAACUGGGUUAGUUGCAUAUAUCUCGCAGCCGCAAGCUGCGAGCCAAACGAUUAACGUUGAGUAACCCAAAUUGAUGUCACCACCCAUCGACCGCAGCUAACAGCCCACCGC ..........((((((((((((......(((((((.(....))))))))...((((.....))))............((((((........))))))..)))))))...))))).... ( -35.80) >DroEre_CAF1 7285 118 + 1 CUUUGUAAACUGGGUUAGUUGCAUAUAUCUCGCAGCCGCAAGCUGCGAGCCAAACGAUUAACGUUGAGUAACCCAAAUUGAUGUCACCACCCAUCGACCACAGCCCACAACUAACGGC ...(((....(((((((.((........(((((((.(....))))))))...((((.....)))))).)))))))..((((((........))))))..)))(((..........))) ( -32.70) >DroYak_CAF1 1293 108 + 1 CUUUGUAAACUGGGUUAGUUGCAUAUAUCUCGCAGCCGCAAGCUGCGAGCCAAACGAUUAACGUUGAGUAACCCAAAUUGAUGUCACCACCCAUCGACCAC----------UAACAGC ..((((....(((((((.((........(((((((.(....))))))))...((((.....)))))).)))))))..((((((........))))))....----------..)))). ( -31.70) >consensus CUUUGUAAACUGGGUUAGUUGCAUAUAUCUCGCAGCCGCAAGCUGCGAGCCAAACGAUUAACGUUGAGUAACCCAAAUUGAUGUCACCACCCAUCGACCACAGCUAACAGCCAAC_GC ...(((....(((((((.((........(((((((.(....))))))))...((((.....)))))).)))))))..((((((........))))))..)))................ (-30.77 = -30.77 + 0.00)

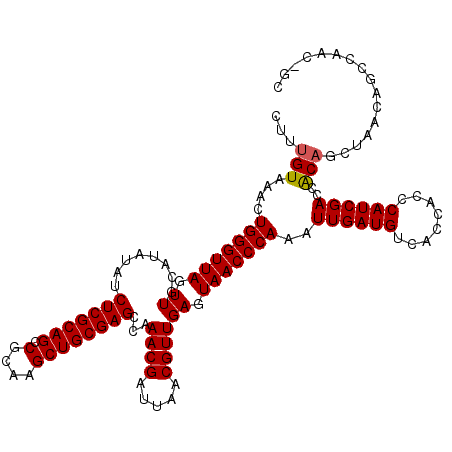

| Location | 18,748,475 – 18,748,587 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 90.54 |
| Mean single sequence MFE | -40.58 |
| Consensus MFE | -34.50 |
| Energy contribution | -34.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.951100 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18748475 112 - 27905053 ------GGCUGUUAGCUGCGGUCGAUGGGUGGUGACAUCAAUUUGGGUUACUCAACGUUAAUCGUUUGGCUCGCAGCUUGCGGCUGCGAGAUAUAUGCAACUAACCCAGUUUACAAAG ------((((((.....))))))....((((....))))...((((((((((.((((.....)))).))((((((((.....))))))))...........))))))))......... ( -41.00) >DroSec_CAF1 7223 118 - 1 GCGGUGGGCUGUUAGCUGCGGUCGAUGGGUGGUGACAUCAAUUUGGGUUACUCAACGUUAAUCGUUUGGCUCGCAGCUUGCGGCUGCGAGAUAUAUGCAACUAACCCAGUUUACAAAG ...((((((((((((.((((((..(((((((....))))...(((((...))))))))..)))......((((((((.....))))))))......))).)))))..))))))).... ( -43.60) >DroEre_CAF1 7285 118 - 1 GCCGUUAGUUGUGGGCUGUGGUCGAUGGGUGGUGACAUCAAUUUGGGUUACUCAACGUUAAUCGUUUGGCUCGCAGCUUGCGGCUGCGAGAUAUAUGCAACUAACCCAGUUUACAAAG ........((((((((((.((((((..((((....))))...)))((((.((.((((.....)))).))((((((((.....))))))))........)))).))))))))))))).. ( -42.20) >DroYak_CAF1 1293 108 - 1 GCUGUUA----------GUGGUCGAUGGGUGGUGACAUCAAUUUGGGUUACUCAACGUUAAUCGUUUGGCUCGCAGCUUGCGGCUGCGAGAUAUAUGCAACUAACCCAGUUUACAAAG .......----------((((.(....((((....))))....(((((((((.((((.....)))).))((((((((.....))))))))...........)))))))).)))).... ( -35.50) >consensus GC_GUUAGCUGUUAGCUGCGGUCGAUGGGUGGUGACAUCAAUUUGGGUUACUCAACGUUAAUCGUUUGGCUCGCAGCUUGCGGCUGCGAGAUAUAUGCAACUAACCCAGUUUACAAAG ....................((.(((.((((....))))....(((((((((.((((.....)))).))((((((((.....))))))))...........)))))))))).)).... (-34.50 = -34.50 + 0.00)

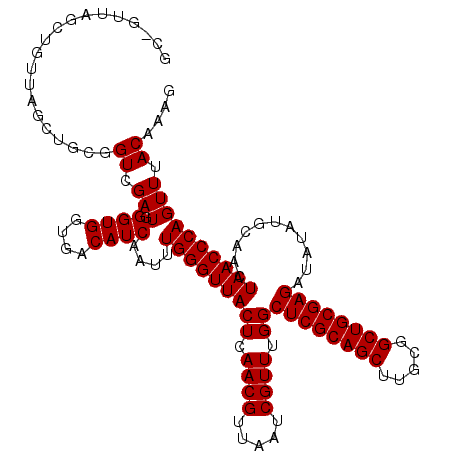

| Location | 18,748,513 – 18,748,620 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 82.63 |
| Mean single sequence MFE | -39.52 |
| Consensus MFE | -29.14 |
| Energy contribution | -29.32 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.831827 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18748513 107 - 27905053 AGCGGGCAAAUGUUGGUGGCGGUGGGUGGUGCG------------GGCUGUUAGCUGCGGUCGAUGGGUGGUGACAUCAAUUUGGGUUACUCAACGUUAAUCGUUUGGCUCGCAGCUUG .(((((((((((....(((((.(((((..(.((------------((((((.....))))))....((((....))))....)).)..))))).)))))..))))).))))))...... ( -41.80) >DroSec_CAF1 7261 119 - 1 AGCGGGCAGAUGUGGGUGAUGUUGGAUGGUGGGUGGUGGGCGGUGGGCUGUUAGCUGCGGUCGAUGGGUGGUGACAUCAAUUUGGGUUACUCAACGUUAAUCGUUUGGCUCGCAGCUUG .(((((((((((....(((((((((((.(..(.(((..(.(....).)..))).)..).)))....((((....)))).............))))))))..))))).))))))...... ( -44.00) >DroEre_CAF1 7323 119 - 1 AGCGGGCAAAUGUGGGUGUUUGUGGGUGGUGGGCGGUGGGCCGUUAGUUGUGGGCUGUGGUCGAUGGGUGGUGACAUCAAUUUGGGUUACUCAACGUUAAUCGUUUGGCUCGCAGCUUG .((..((((((......))))))..))....(((((....)))))((((((((((((((((..(((((((....))))...(((((...))))))))..))))..)))))))))))).. ( -39.30) >DroYak_CAF1 1331 109 - 1 AGUGGGCAAAUGUGGGUGGUGCUGGGUGGUGGGCUGUUAGCUGUUA----------GUGGUCGAUGGGUGGUGACAUCAAUUUGGGUUACUCAACGUUAAUCGUUUGGCUCGCAGCUUG .(((((((((((....(..((.((((((((.(((..((((...)))----------)..))).)).((((....)))).........)))))).))..)..))))).))))))...... ( -33.00) >consensus AGCGGGCAAAUGUGGGUGGUGGUGGGUGGUGGGCGGUGGGC_GUUAGCUGUUAGCUGCGGUCGAUGGGUGGUGACAUCAAUUUGGGUUACUCAACGUUAAUCGUUUGGCUCGCAGCUUG .(((((((((((....(((((.(((((((................(((.....)))....((((..((((....))))...)))).))))))).)))))..))))).))))))...... (-29.14 = -29.32 + 0.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:37:46 2006