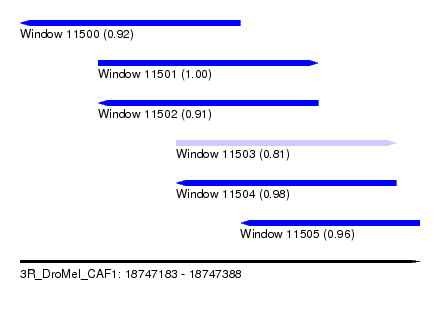
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,747,183 – 18,747,388 |
| Length | 205 |
| Max. P | 0.995717 |

| Location | 18,747,183 – 18,747,296 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.85 |
| Mean single sequence MFE | -30.87 |
| Consensus MFE | -28.15 |
| Energy contribution | -28.40 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.916129 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18747183 113 - 27905053 GGAAAAGGA-------AAAACUCUGGCCUUAACAGGUCGCCCUGAAAAUUAAAAGUCCGCCGCGAAUGAAGCGUUAAUGAUGGCGAGUAAGUAAACAGUUUUUGGGUCGACAGCAUAAAU ((...(((.-------.....)))..)).......((((.((..((((((.......((((((.((((...))))...).))))).((......))))))))..)).))))......... ( -23.60) >DroSec_CAF1 5967 113 - 1 GGAAAAGGA-------AAAACCCUGGCCUUAACAGGUCGCCCUGAAAAUUAAAAGUCCGCCGCGAAUGAAGCGUUAAUGAAGGCGAGUAAGUAAACAGUUUUUGGGGCGACAGCAUAAAU ((...(((.-------.....)))..)).......(((((((..((((((.......((((...((((...))))......)))).((......))))))))..)))))))......... ( -33.30) >DroSim_CAF1 5981 113 - 1 GGAAAAGGA-------AAAACCCUGGCCUUAACAGGUCGCCCUGAAAAUUAAAAGUCCGCCGCGAAUGAAGCGUUAAUGAAGGCGAGUAAGUAAACAGUUUUUGGGGCGACAGCAUAAAU ((...(((.-------.....)))..)).......(((((((..((((((.......((((...((((...))))......)))).((......))))))))..)))))))......... ( -33.30) >DroEre_CAF1 5990 120 - 1 GGAAAAUGGAAAGAGGAAAACCCUGGCCUUAACAGGUCGCCCUGAAAAUUAAAAGUCCGCCGCGAAUGAAGCGUUAAUGAAGGCGAGUAAGUAAACAGUUUUUGGGGCGACAGCAUAAAU .......((..((.((....))))..)).......(((((((..((((((.......((((...((((...))))......)))).((......))))))))..)))))))......... ( -33.30) >consensus GGAAAAGGA_______AAAACCCUGGCCUUAACAGGUCGCCCUGAAAAUUAAAAGUCCGCCGCGAAUGAAGCGUUAAUGAAGGCGAGUAAGUAAACAGUUUUUGGGGCGACAGCAUAAAU ......................(((.......)))(((((((..((((((.......((((...((((...))))......)))).((......))))))))..)))))))......... (-28.15 = -28.40 + 0.25)

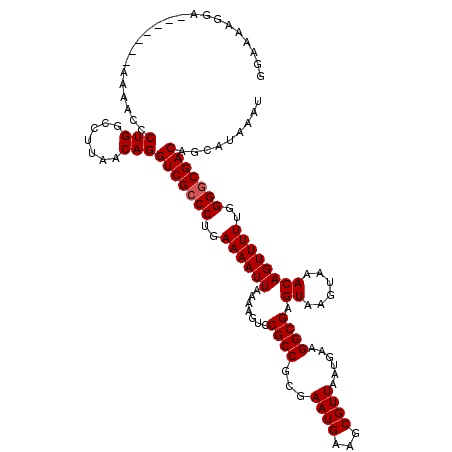
| Location | 18,747,223 – 18,747,336 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.55 |
| Mean single sequence MFE | -39.00 |
| Consensus MFE | -30.95 |
| Energy contribution | -32.13 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.14 |
| Mean z-score | -3.44 |
| Structure conservation index | 0.79 |
| SVM decision value | 2.61 |
| SVM RNA-class probability | 0.995717 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18747223 113 + 27905053 UCAUUAACGCUUCAUUCGCGGCGGACUUUUAAUUUUCAGGGCGACCUGUUAAGGCCAGAGUUUU-------UCCUUUUCCUUUGGCCGGGGCAUCGCCAAAAAGCCCUAUAACCGUGAAU .............((((((((...........((..((((....))))..))((((((((....-------........))))))))(((((...........)))))....)))))))) ( -36.50) >DroSec_CAF1 6007 113 + 1 UCAUUAACGCUUCAUUCGCGGCGGACUUUUAAUUUUCAGGGCGACCUGUUAAGGCCAGGGUUUU-------UCCUUUUCCUUUGGCCGGGGCAUCGCCAAAGAGCCUCAUAGCCGUGAAU .............(((((((((..........((..((((....))))..))((((((((....-------........))))))))(((((.((......)))))))...))))))))) ( -42.40) >DroSim_CAF1 6021 113 + 1 UCAUUAACGCUUCAUUCGCGGCGGACUUUUAAUUUUCAGGGCGACCUGUUAAGGCCAGGGUUUU-------UCCUUUUCCUUUGGCCGGGGCAUCGCCAAAGAGCCUUAUAGCCGUGAAU .............(((((((((..............((((....)))).((((((.((((....-------))))....(((((((.((....))))))))).))))))..))))))))) ( -40.50) >DroEre_CAF1 6030 101 + 1 UCAUUAACGCUUCAUUCGCGGCGGACUUUUAAUUUUCAGGGCGACCUGUUAAGGCCAGGGUUUUCCUCUUUCCAUUUUCCUUUGG-------------------CCUUAUAGCCGCGAAU .............(((((((((..............((((....)))).(((((((((((...................))))))-------------------)))))..))))))))) ( -36.61) >consensus UCAUUAACGCUUCAUUCGCGGCGGACUUUUAAUUUUCAGGGCGACCUGUUAAGGCCAGGGUUUU_______UCCUUUUCCUUUGGCCGGGGCAUCGCCAAAGAGCCUUAUAGCCGUGAAU .............(((((((((..........((..((((....))))..))((((((((...................))))))))(((((...........)))))...))))))))) (-30.95 = -32.13 + 1.19)


| Location | 18,747,223 – 18,747,336 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.55 |
| Mean single sequence MFE | -34.15 |
| Consensus MFE | -24.21 |
| Energy contribution | -25.64 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.909676 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18747223 113 - 27905053 AUUCACGGUUAUAGGGCUUUUUGGCGAUGCCCCGGCCAAAGGAAAAGGA-------AAAACUCUGGCCUUAACAGGUCGCCCUGAAAAUUAAAAGUCCGCCGCGAAUGAAGCGUUAAUGA ((((.((((....((((((((.((((((.....(((((.((........-------....)).))))).......)))))).........)))))))))))).))))............. ( -32.40) >DroSec_CAF1 6007 113 - 1 AUUCACGGCUAUGAGGCUCUUUGGCGAUGCCCCGGCCAAAGGAAAAGGA-------AAAACCCUGGCCUUAACAGGUCGCCCUGAAAAUUAAAAGUCCGCCGCGAAUGAAGCGUUAAUGA ((((.((((..((((((((((((((.........)))))))....(((.-------.....)))))))))).((((....))))..............)))).))))............. ( -35.80) >DroSim_CAF1 6021 113 - 1 AUUCACGGCUAUAAGGCUCUUUGGCGAUGCCCCGGCCAAAGGAAAAGGA-------AAAACCCUGGCCUUAACAGGUCGCCCUGAAAAUUAAAAGUCCGCCGCGAAUGAAGCGUUAAUGA ((((.((((..((((((((((((((.........)))))))....(((.-------.....)))))))))).((((....))))..............)))).))))............. ( -35.70) >DroEre_CAF1 6030 101 - 1 AUUCGCGGCUAUAAGG-------------------CCAAAGGAAAAUGGAAAGAGGAAAACCCUGGCCUUAACAGGUCGCCCUGAAAAUUAAAAGUCCGCCGCGAAUGAAGCGUUAAUGA (((((((((..(((((-------------------((.........(....).(((.....)))))))))).((((....))))..............)))))))))............. ( -32.70) >consensus AUUCACGGCUAUAAGGCUCUUUGGCGAUGCCCCGGCCAAAGGAAAAGGA_______AAAACCCUGGCCUUAACAGGUCGCCCUGAAAAUUAAAAGUCCGCCGCGAAUGAAGCGUUAAUGA ((((.((((..((((((((((((((.........)))))))....(((.............)))))))))).((((....))))..............)))).))))............. (-24.21 = -25.64 + 1.44)


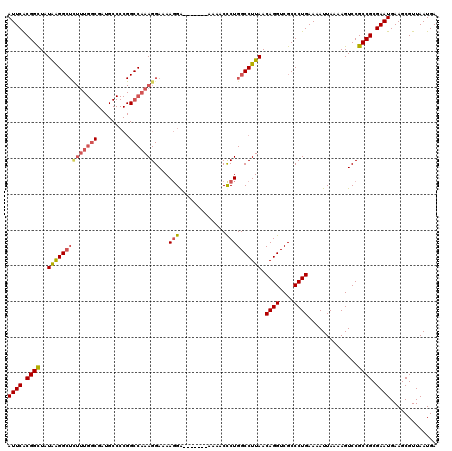
| Location | 18,747,263 – 18,747,376 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 88.44 |
| Mean single sequence MFE | -37.40 |
| Consensus MFE | -26.81 |
| Energy contribution | -27.25 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.805779 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
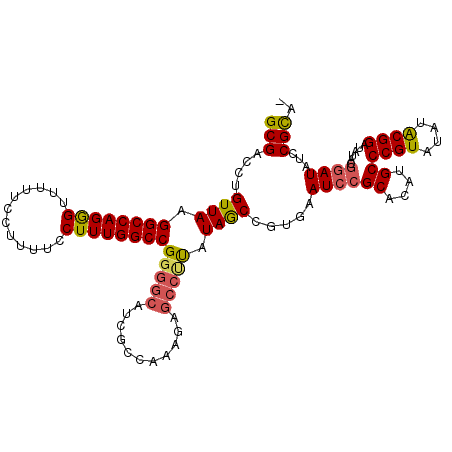
>3R_DroMel_CAF1 18747263 113 + 27905053 GCGACCUGUUAAGGCCAGAGUUUUUCCUUUUCCUUUGGCCGGGGCAUCGCCAAAAAGCCCUAUAACCGUGAAUCCGCACAUGCCCGUAUAUACGGAUAUACGGAUAUACGUAU .......((((.((((((((............))))))))(((((...........))))).))))((((.(((((...(((.((((....)))).))).))))).))))... ( -35.40) >DroSec_CAF1 6047 112 + 1 GCGACCUGUUAAGGCCAGGGUUUUUCCUUUUCCUUUGGCCGGGGCAUCGCCAAAGAGCCUCAUAGCCGUGAAUCCGCACAUGCCCGUAUAUACGGAUAUACGGAUAUCCGCA- (((..((((...((((((((............))))))))(((((.((......)))))))))))(((((.(((((...(((....)))...))))).))))).....))).- ( -39.10) >DroSim_CAF1 6061 112 + 1 GCGACCUGUUAAGGCCAGGGUUUUUCCUUUUCCUUUGGCCGGGGCAUCGCCAAAGAGCCUUAUAGCCGUGAAUCCGCACAUGCCCGUAUAUACGGAUAUACGGAUAUCCGCA- (((..((((.(((((.((((....))))....(((((((.((....))))))))).)))))))))(((((.(((((...(((....)))...))))).))))).....))).- ( -37.90) >DroYak_CAF1 13 97 + 1 GCGAGCUGUUAAGGCCAGGGCUUUUCCUUUUCCUUUGGCCG-------GCCAAAGAGCCUUAUAGCCGUGAAUCCGCACAUGCCCGUAUAUGCGG--------AUAUCCGCA- (((.(((((.(((((.((((....))))....((((((...-------.)))))).)))))))))).....(((((((.(((....))).)))))--------))...))).- ( -37.20) >consensus GCGACCUGUUAAGGCCAGGGUUUUUCCUUUUCCUUUGGCCGGGGCAUCGCCAAAGAGCCUUAUAGCCGUGAAUCCGCACAUGCCCGUAUAUACGGAUAUACGGAUAUCCGCA_ (((....((((.((((((((............))))))))(((((...........))))).)))).....((((((....))((((....))))......))))...))).. (-26.81 = -27.25 + 0.44)


| Location | 18,747,263 – 18,747,376 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 88.44 |
| Mean single sequence MFE | -40.08 |
| Consensus MFE | -33.94 |
| Energy contribution | -34.75 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.976996 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
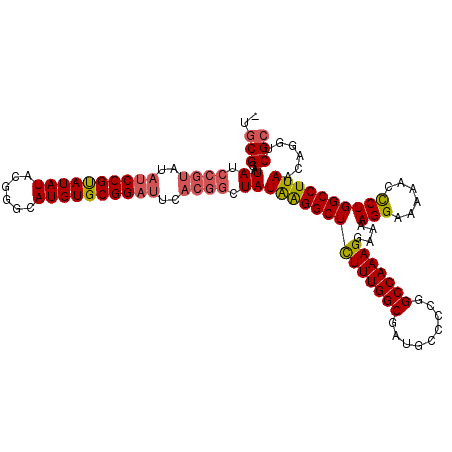
>3R_DroMel_CAF1 18747263 113 - 27905053 AUACGUAUAUCCGUAUAUCCGUAUAUACGGGCAUGUGCGGAUUCACGGUUAUAGGGCUUUUUGGCGAUGCCCCGGCCAAAGGAAAAGGAAAAACUCUGGCCUUAACAGGUCGC ...(((..((((((((((((((....))))..))))))))))..)))((((..((((...........)))).(((((.((............)).))))).))))....... ( -38.10) >DroSec_CAF1 6047 112 - 1 -UGCGGAUAUCCGUAUAUCCGUAUAUACGGGCAUGUGCGGAUUCACGGCUAUGAGGCUCUUUGGCGAUGCCCCGGCCAAAGGAAAAGGAAAAACCCUGGCCUUAACAGGUCGC -....((.((((((((((((((....))))..)))))))))))).(((((.((((((((((((((.........)))))))....(((......))))))))))...))))). ( -41.90) >DroSim_CAF1 6061 112 - 1 -UGCGGAUAUCCGUAUAUCCGUAUAUACGGGCAUGUGCGGAUUCACGGCUAUAAGGCUCUUUGGCGAUGCCCCGGCCAAAGGAAAAGGAAAAACCCUGGCCUUAACAGGUCGC -....((.((((((((((((((....))))..)))))))))))).(((((.((((((((((((((.........)))))))....(((......))))))))))...))))). ( -41.80) >DroYak_CAF1 13 97 - 1 -UGCGGAUAU--------CCGCAUAUACGGGCAUGUGCGGAUUCACGGCUAUAAGGCUCUUUGGC-------CGGCCAAAGGAAAAGGAAAAGCCCUGGCCUUAACAGCUCGC -.(((((.((--------((((((((......))))))))))))..((((.(((((((((((((.-------...))))))....(((......))))))))))..))))))) ( -38.50) >consensus _UGCGGAUAUCCGUAUAUCCGUAUAUACGGGCAUGUGCGGAUUCACGGCUAUAAGGCUCUUUGGCGAUGCCCCGGCCAAAGGAAAAGGAAAAACCCUGGCCUUAACAGGUCGC ..(((..((.((((..((((((((((......))))))))))..)))).))((((((((((((((.........)))))))....(((......))))))))))......))) (-33.94 = -34.75 + 0.81)

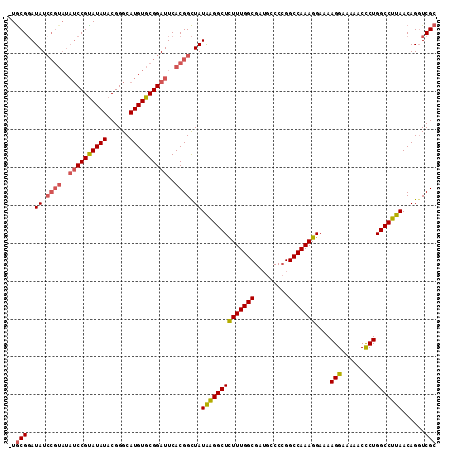
| Location | 18,747,296 – 18,747,388 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 85.71 |
| Mean single sequence MFE | -36.70 |
| Consensus MFE | -23.50 |
| Energy contribution | -25.00 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.19 |
| Mean z-score | -3.66 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.960069 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18747296 92 - 27905053 AUGUGCGGAUAUAUACGUAUAUCCGUAUAUCCGUAUAUACGGGCAUGUGCGGAUUCACGGUUAUAGGGCUUUUUGGCGAUGCCCCGGCCAAA .((((((((((((....))))))))))))(((((((((......))))))))).....((((...((((...........)))).))))... ( -34.90) >DroSec_CAF1 6080 90 - 1 AUGUGCGGAUG--UGCGGAUAUCCGUAUAUCCGUAUAUACGGGCAUGUGCGGAUUCACGGCUAUGAGGCUCUUUGGCGAUGCCCCGGCCAAA (((((((((((--(((((....))))))))))))))))..((((((.(((.((.....((((....))))..)).)))))))))........ ( -41.50) >DroSim_CAF1 6094 90 - 1 AUGUGCGGAUG--UGCGGAUAUCCGUAUAUCCGUAUAUACGGGCAUGUGCGGAUUCACGGCUAUAAGGCUCUUUGGCGAUGCCCCGGCCAAA (((((((((((--(((((....))))))))))))))))..((((((.(((.((.....((((....))))..)).)))))))))........ ( -41.50) >DroYak_CAF1 46 75 - 1 AAGUGCGGAUG--UGCGGAUAU--------CCGCAUAUACGGGCAUGUGCGGAUUCACGGCUAUAAGGCUCUUUGGC-------CGGCCAAA ..((.((.(((--(((((....--------)))))))).)).))..(((......)))((((....((((....)))-------)))))... ( -28.90) >consensus AUGUGCGGAUG__UGCGGAUAUCCGUAUAUCCGUAUAUACGGGCAUGUGCGGAUUCACGGCUAUAAGGCUCUUUGGCGAUGCCCCGGCCAAA (((((((((((..(((((....))))))))))))))))..((((.(((((.(.....).)).)))..))))((((((.........)))))) (-23.50 = -25.00 + 1.50)

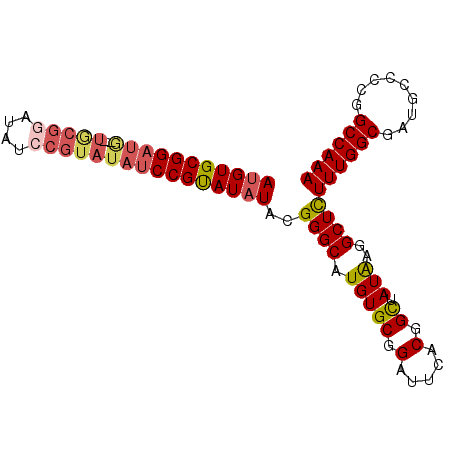

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:37:40 2006