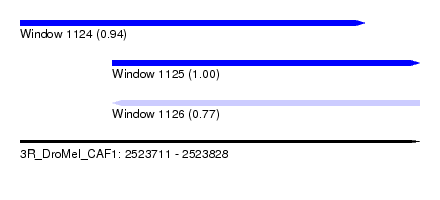
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,523,711 – 2,523,828 |
| Length | 117 |
| Max. P | 0.998165 |

| Location | 2,523,711 – 2,523,812 |
|---|---|
| Length | 101 |
| Sequences | 3 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 76.95 |
| Mean single sequence MFE | -21.07 |
| Consensus MFE | -15.32 |
| Energy contribution | -15.43 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.939439 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2523711 101 + 27905053 ------GGACUUAAUUGCGUCCGAAAGAGUAAUGAUAGCAUUUCUUUGGUAGUCUGGAGAUAUGUUUGAAUAUGUAAAUAUUAUUAAAUAUUUCUAGAUGUAUUUGC ------((((........))))...((((.((((....)))).))))....((((((((((((.....(((((....))))).....))))))))))))........ ( -24.40) >DroSec_CAF1 53063 84 + 1 A---------------GCGUCCGAAAGAGUAAUGAUAGCAUUUCUUUGGUAGUCUGGAGGUAUGUUUGAAUAAGU--------UUAAAUAUUUCUAGAUGUAUUUAC .---------------((..((.(((((..((((....)))))))))))..((((((((..(((((((((....)--------))))))))))))))))))...... ( -16.90) >DroSim_CAF1 51994 94 + 1 UAUGGUGGACUUAAUUGCGUCCGAAAGACUAAUGAUAGCAUUUCUUUGGUUGGCUGGAGAUAUGUU-----AUGU--------UUAAAUAUUUCUAGAUGUAUUUAC ...........(((.((((((.((((...(((..(((((((.((((..(....)..)))).)))))-----))..--------)))....))))..)))))).))). ( -21.90) >consensus ______GGACUUAAUUGCGUCCGAAAGAGUAAUGAUAGCAUUUCUUUGGUAGUCUGGAGAUAUGUUUGAAUAUGU________UUAAAUAUUUCUAGAUGUAUUUAC ...............(((((((....).........(((((.((((..(....)..)))).)))))..............................))))))..... (-15.32 = -15.43 + 0.11)

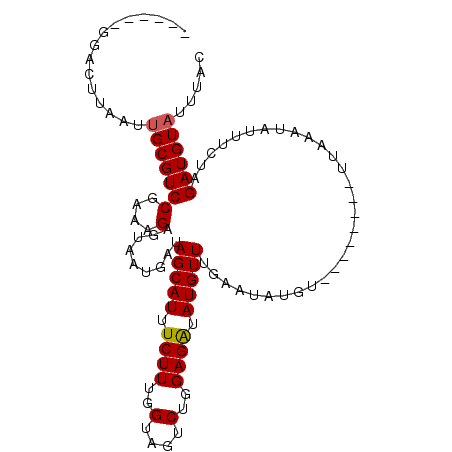
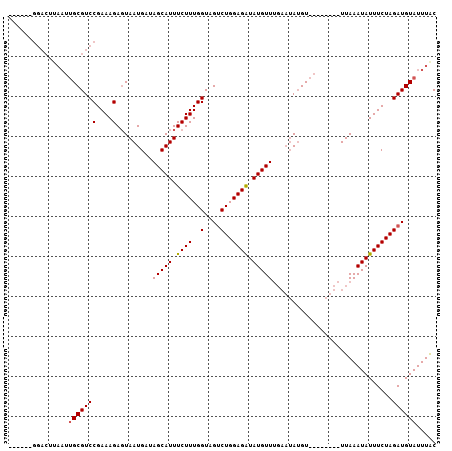
| Location | 2,523,738 – 2,523,828 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 80.81 |
| Mean single sequence MFE | -22.13 |
| Consensus MFE | -18.87 |
| Energy contribution | -18.43 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.90 |
| Structure conservation index | 0.85 |
| SVM decision value | 3.02 |
| SVM RNA-class probability | 0.998165 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2523738 90 + 27905053 GAUAGCAUUUCUUUGGUAGUCUGGAGAUAUGUUUGAAUAUGUAAAUAUUAUUAAAUAUUUCUAGAUGUAUUUGCAUUUAAGAAAUGGCGU---- .....((((((((..(((((((((((((((.....(((((....))))).....)))))))))))).....)))....))))))))....---- ( -23.80) >DroSec_CAF1 53081 86 + 1 GAUAGCAUUUCUUUGGUAGUCUGGAGGUAUGUUUGAAUAAGU--------UUAAAUAUUUCUAGAUGUAUUUACAUUUAAGAAAUGACGUAGAA ....(((((((((..(((((((((((..(((((((((....)--------)))))))))))))))).....)))....)))))))).)...... ( -20.20) >DroSim_CAF1 52027 75 + 1 GAUAGCAUUUCUUUGGUUGGCUGGAGAUAUGUU-----AUGU--------UUAAAUAUUUCUAGAUGUAUUUACAUUUAAGAAAUGA------A .(((((((.((((..(....)..)))).)))))-----))..--------.....(((((((((((((....)))))).))))))).------. ( -22.40) >consensus GAUAGCAUUUCUUUGGUAGUCUGGAGAUAUGUUUGAAUAUGU________UUAAAUAUUUCUAGAUGUAUUUACAUUUAAGAAAUGACGU___A ...(((((.((((..(....)..)))).)))))......................(((((((((((((....)))))).)))))))........ (-18.87 = -18.43 + -0.44)

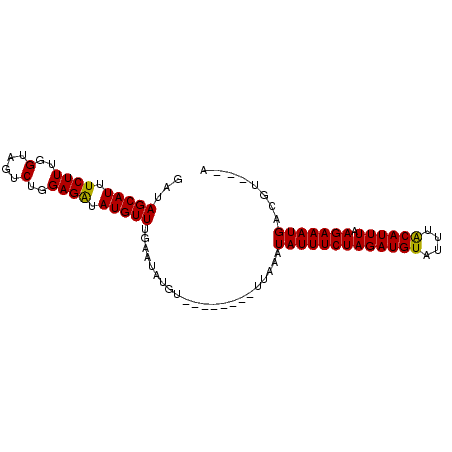
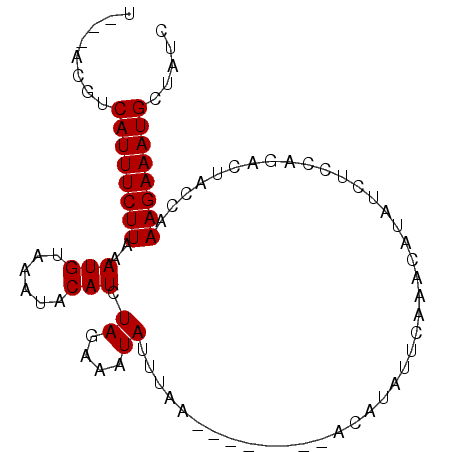
| Location | 2,523,738 – 2,523,828 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 80.81 |
| Mean single sequence MFE | -7.37 |
| Consensus MFE | -5.20 |
| Energy contribution | -5.20 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.766795 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2523738 90 - 27905053 ----ACGCCAUUUCUUAAAUGCAAAUACAUCUAGAAAUAUUUAAUAAUAUUUACAUAUUCAAACAUAUCUCCAGACUACCAAAGAAAUGCUAUC ----..((.(((((((...((......))(((.((.((((.....(((((....))))).....)))).)).)))......))))))))).... ( -8.50) >DroSec_CAF1 53081 86 - 1 UUCUACGUCAUUUCUUAAAUGUAAAUACAUCUAGAAAUAUUUAA--------ACUUAUUCAAACAUACCUCCAGACUACCAAAGAAAUGCUAUC ......(.((((((((..((((....))))...(((.((.....--------...))))).....................))))))))).... ( -6.90) >DroSim_CAF1 52027 75 - 1 U------UCAUUUCUUAAAUGUAAAUACAUCUAGAAAUAUUUAA--------ACAU-----AACAUAUCUCCAGCCAACCAAAGAAAUGCUAUC .------.((((((((..((((....))))((.((.(((((((.--------...)-----)).)))).)).)).......))))))))..... ( -6.70) >consensus U___ACGUCAUUUCUUAAAUGUAAAUACAUCUAGAAAUAUUUAA________ACAUAUUCAAACAUAUCUCCAGACUACCAAAGAAAUGCUAUC ........((((((((..(((......))).((....))..........................................))))))))..... ( -5.20 = -5.20 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:49:59 2006