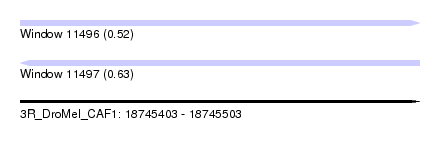
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,745,403 – 18,745,503 |
| Length | 100 |
| Max. P | 0.626812 |

| Location | 18,745,403 – 18,745,503 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 90.83 |
| Mean single sequence MFE | -29.93 |
| Consensus MFE | -23.87 |
| Energy contribution | -24.18 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.80 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.522523 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18745403 100 + 27905053 CCCAUAUAAAUUCGUAUAUUAGCAUAC---------ACAUUUGCAUGCCGCGUCCCCUGGGCGAUUGCCAUGCGAUAAUUAUGGCAGUCGAAUCCCGAAAAGGACGCAG ..(((.(((((..((((......))))---------..))))).)))..((((((..(((((((((((((((.......)))))))))))...))))....)))))).. ( -34.10) >DroSec_CAF1 4203 109 + 1 CCCAUAUAAAUUCGUAUAUUAGCAUACACAGCAGACACAUUUGCACGCCGCGUCCCCUGGGCGAUUGCCAUGCGAUAAUUAUGGCAGGCGAAUACCGAAAAGGACGCCG .(((((.....((((((....(((......(((((....))))).((((.((.....))))))..))).))))))....)))))..((((....((.....)).)))). ( -29.90) >DroSim_CAF1 4216 109 + 1 CCCAUAUAAAUUCGUAUAUUAGCAUACACAGCAGACACAUUUGCACGCCGCGUCCCCUGGGCGAUUGCCAUGCGAUAAUUAUGGCAGGCGAAUCCCGAAAAGCACGCAG .....................((.......(((((....))))).((((.(((((...)))))..(((((((.......)))))))))))...............)).. ( -27.60) >DroEre_CAF1 4216 108 + 1 CCCAUAUAAAUUCGUAUAUUAGCAUACACAGAAUACACAUUUGCAGGCUGCGUCC-CUGGGCGAUUGCUAUGCGAUAAUUAUGGCAGGCGUAUCCCGAAAAGGACGCAG .(((((((......)))))..(((.................))).))((((((((-.((((((.((((((((.......)))))))).))...))))....)))))))) ( -28.13) >consensus CCCAUAUAAAUUCGUAUAUUAGCAUACACAGCAGACACAUUUGCACGCCGCGUCCCCUGGGCGAUUGCCAUGCGAUAAUUAUGGCAGGCGAAUCCCGAAAAGGACGCAG .....................(((.................))).....((((((..((((((.((((((((.......)))))))).))...))))....)))))).. (-23.87 = -24.18 + 0.31)

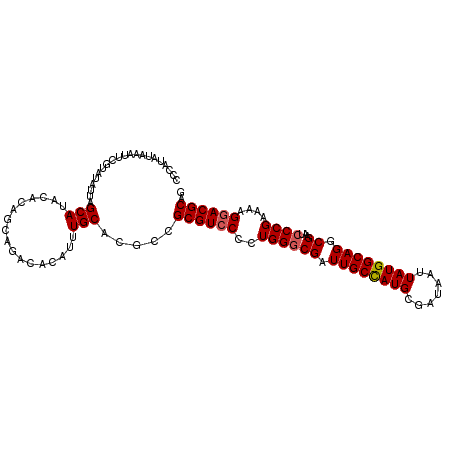
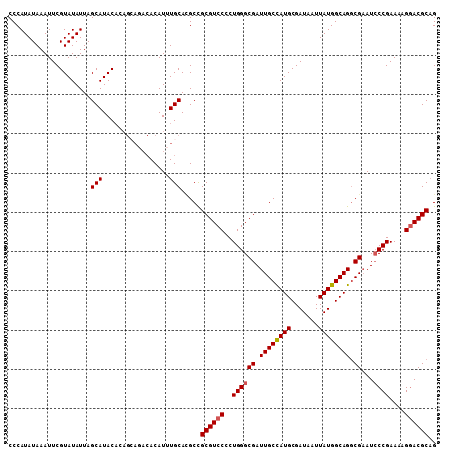
| Location | 18,745,403 – 18,745,503 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 90.83 |
| Mean single sequence MFE | -34.65 |
| Consensus MFE | -26.86 |
| Energy contribution | -28.30 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.626812 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18745403 100 - 27905053 CUGCGUCCUUUUCGGGAUUCGACUGCCAUAAUUAUCGCAUGGCAAUCGCCCAGGGGACGCGGCAUGCAAAUGU---------GUAUGCUAAUAUACGAAUUUAUAUGGG ..(((((((((..(((...(((.((((((.........)))))).)))))))))))))))((((((((....)---------))))))).(((((......)))))... ( -37.30) >DroSec_CAF1 4203 109 - 1 CGGCGUCCUUUUCGGUAUUCGCCUGCCAUAAUUAUCGCAUGGCAAUCGCCCAGGGGACGCGGCGUGCAAAUGUGUCUGCUGUGUAUGCUAAUAUACGAAUUUAUAUGGG (((((((((((..((((((.((((((.((....)).))).)))))).))).)))))))))((((((((...((....))..))))))))......))............ ( -36.10) >DroSim_CAF1 4216 109 - 1 CUGCGUGCUUUUCGGGAUUCGCCUGCCAUAAUUAUCGCAUGGCAAUCGCCCAGGGGACGCGGCGUGCAAAUGUGUCUGCUGUGUAUGCUAAUAUACGAAUUUAUAUGGG ..((((.((((..((((((.((((((.((....)).))).))))))).)).)))).))))((((((((...((....))..)))))))).(((((......)))))... ( -33.00) >DroEre_CAF1 4216 108 - 1 CUGCGUCCUUUUCGGGAUACGCCUGCCAUAAUUAUCGCAUAGCAAUCGCCCAG-GGACGCAGCCUGCAAAUGUGUAUUCUGUGUAUGCUAAUAUACGAAUUUAUAUGGG ((((((((((...(((....((.(((.((....)).)))..)).....)))))-))))))))(((....(((((.((((.((((((....)))))))))).)))))))) ( -32.20) >consensus CUGCGUCCUUUUCGGGAUUCGCCUGCCAUAAUUAUCGCAUGGCAAUCGCCCAGGGGACGCGGCGUGCAAAUGUGUCUGCUGUGUAUGCUAAUAUACGAAUUUAUAUGGG ..(((((((((..(((...((..((((((.........))))))..))))))))))))))((((((((...(......)..)))))))).(((((......)))))... (-26.86 = -28.30 + 1.44)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:37:33 2006