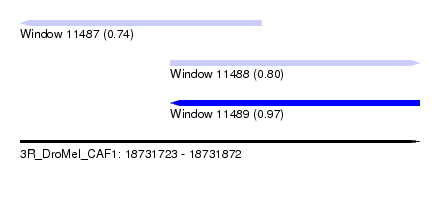
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,731,723 – 18,731,872 |
| Length | 149 |
| Max. P | 0.972541 |

| Location | 18,731,723 – 18,731,813 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 78.63 |
| Mean single sequence MFE | -29.66 |
| Consensus MFE | -18.71 |
| Energy contribution | -19.32 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.736804 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18731723 90 - 27905053 AAAUGCUCGCCGGCAGCAGGAAUCUCCUGCCAUC----------------CAGUCCA--------CCCAGUCAUCUGAAAUGAAGGCUCUUUUGCCGGAGGCUGCAGUAAUUAC ...(((..(((((..(((((.....)))))...)----------------)..(((.--------..(((....))).......(((......))))))))).)))........ ( -29.10) >DroSec_CAF1 165313 97 - 1 AAAUGCUCGCCGGCAGCAGGAAUCUCCUGCCAUC----------------CUG-CCAUCCAGUCACCCAGUCAUCCGAAAUGAAGGCUCUUUCGCCGGAGGCUGCAGUAAUUAC ...(((..((((((((((((.....)))))....----------------.))-)).(((..........((((.....)))).(((......))))))))).)))........ ( -31.10) >DroSim_CAF1 156994 89 - 1 AAAUGCUCGCCGGCAGCAGGAAUCUCCUGCCAUC----------------CUG-CCAUCCA--------GUCAUCCGAAAUGAAGGCUCUUUCGCCGGAGGCUGCAGUAAUUAC ...(((..((((((((((((.....)))))....----------------.))-)).(((.--------.((((.....)))).(((......))))))))).)))........ ( -31.10) >DroEre_CAF1 163047 85 - 1 ------------GGAGCAGGAAUCUGCUGCCAUC----------------CAG-UCACCCAGUCAUCCAGUCGUCUGAAAUGAAGGCUCCUUCGCCGGAGGCUGCAGUAAUUAC ------------(((((((....))))).))...----------------(((-((.((...((((.(((....)))..)))).(((......))))).))))).......... ( -24.90) >DroYak_CAF1 163950 113 - 1 AAAUGCUCGCCGGCAGCAGGAAUCUCCUGCCAUCCUGCCAUACAGUCACCCAG-UCAACCAGUCAUCCAGUCAUCUGAAAUGAAGGCUCCUUCGCCGGAGGCUGCAGUAAUUAC ...(((..((((((((((((.....))))).....))))..............-....((..((((.(((....)))..)))).(((......))))).))).)))........ ( -32.10) >consensus AAAUGCUCGCCGGCAGCAGGAAUCUCCUGCCAUC________________CAG_CCAUCCAGUCACCCAGUCAUCUGAAAUGAAGGCUCUUUCGCCGGAGGCUGCAGUAAUUAC ...((((....))))(((((.....))))).....................................(((((.((((...(((((....))))).))))))))).......... (-18.71 = -19.32 + 0.61)

| Location | 18,731,779 – 18,731,872 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 87.17 |
| Mean single sequence MFE | -32.92 |
| Consensus MFE | -26.17 |
| Energy contribution | -26.55 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.802253 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18731779 93 + 27905053 ------GAUGGCAGGAGAUUCCUGCUGCCGGCGAGCAUUUAACCUGCUCCUCUUCCUGUCGCCAGUAGAGUCC-UUGCCGCCAC-------UGUCUCCAUGCUGCUC ------...((((((((((....(.((.(((((((((.......)))))((((..(((....))).))))...-..)))).)))-------.)))))).)))).... ( -31.70) >DroSec_CAF1 165376 90 + 1 ------GAUGGCAGGAGAUUCCUGCUGCCGGCGAGCAUUUAACCUGCUCCUCUUCCUGUCGCCAGAAGAGUCC-UUGC---CAC-------UAUCUCCAUGCCGCUC ------...((((((((((..(....)..((((((((.......)))))((((((.((....))))))))...-..))---)..-------.)))))).)))).... ( -30.30) >DroSim_CAF1 157049 93 + 1 ------GAUGGCAGGAGAUUCCUGCUGCCGGCGAGCAUUUAACCUGCUCCUCUUCCUGUCGCCAGAAGAGUCC-UUGCCGCCAC-------UGUAUCCAUGCUGCUC ------...(((((.((....)).)))))((((((((.......)))))((((((.((....))))))))...-..)))((...-------.(((....))).)).. ( -30.00) >DroYak_CAF1 164023 107 + 1 UGGCAGGAUGGCAGGAGAUUCCUGCUGCCGGCGAGCAUUUAACCUGCUCCUCUUCCUGUCGCCAGAAGAGUCCUUUGCUGCCUCUGUCCCCUGUCGCCGUUCUGCUC .((((((((((((((.(((....((.((.((.(((((.......)))))((((((.((....)))))))).))...)).))....))).)))...))))))))))). ( -39.70) >consensus ______GAUGGCAGGAGAUUCCUGCUGCCGGCGAGCAUUUAACCUGCUCCUCUUCCUGUCGCCAGAAGAGUCC_UUGCCGCCAC_______UGUCUCCAUGCUGCUC .........(((((.((....)).)))))((((((((.......)))))((((((.((....))))))))......)))............................ (-26.17 = -26.55 + 0.38)

| Location | 18,731,779 – 18,731,872 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 87.17 |
| Mean single sequence MFE | -38.75 |
| Consensus MFE | -30.58 |
| Energy contribution | -31.58 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.972541 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18731779 93 - 27905053 GAGCAGCAUGGAGACA-------GUGGCGGCAA-GGACUCUACUGGCGACAGGAAGAGGAGCAGGUUAAAUGCUCGCCGGCAGCAGGAAUCUCCUGCCAUC------ ..((.((.((....))-------...))(((..-...((((.(((....)))..))))(((((.......)))))))).)).(((((.....)))))....------ ( -38.90) >DroSec_CAF1 165376 90 - 1 GAGCGGCAUGGAGAUA-------GUG---GCAA-GGACUCUUCUGGCGACAGGAAGAGGAGCAGGUUAAAUGCUCGCCGGCAGCAGGAAUCUCCUGCCAUC------ ....((((.((((((.-------...---((..-((.((((((((....)).))))))(((((.......))))).))....))....))))))))))...------ ( -37.30) >DroSim_CAF1 157049 93 - 1 GAGCAGCAUGGAUACA-------GUGGCGGCAA-GGACUCUUCUGGCGACAGGAAGAGGAGCAGGUUAAAUGCUCGCCGGCAGCAGGAAUCUCCUGCCAUC------ ..((.((.((....))-------...))(((..-...((((((((....)).))))))(((((.......)))))))).)).(((((.....)))))....------ ( -37.20) >DroYak_CAF1 164023 107 - 1 GAGCAGAACGGCGACAGGGGACAGAGGCAGCAAAGGACUCUUCUGGCGACAGGAAGAGGAGCAGGUUAAAUGCUCGCCGGCAGCAGGAAUCUCCUGCCAUCCUGCCA (.((((...(((...((((((.....((.((...((.((((((((....)).))))))(((((.......))))).)).)).)).....)))))))))...))))). ( -41.60) >consensus GAGCAGCAUGGAGACA_______GUGGCGGCAA_GGACUCUUCUGGCGACAGGAAGAGGAGCAGGUUAAAUGCUCGCCGGCAGCAGGAAUCUCCUGCCAUC______ .........................(.((((......((((((((....)).))))))(((((.......))))))))).).(((((.....))))).......... (-30.58 = -31.58 + 1.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:37:26 2006