| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,517,440 – 2,517,544 |
| Length | 104 |
| Max. P | 0.863095 |

| Location | 2,517,440 – 2,517,544 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 78.05 |
| Mean single sequence MFE | -28.13 |
| Consensus MFE | -20.93 |
| Energy contribution | -20.05 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.863095 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2517440 104 - 27905053 CCAAUUAGGUGUCAUCAAUGUCACCAAGUAGCGUAUAAAAUA-AACGGCCCACAAUGGGUCAGACAUUCAGUGUUUGGCUUCGGCUCAAGACAGCA------GUUAAUC--------AA .......((((.((....)).)))).....((..........-...(((((.....))))).........((.((.(((....))).)).)).)).------.......--------.. ( -26.80) >DroVir_CAF1 88964 116 - 1 CCAAUUAGGUGUCAUCAAUGUCAGCGAGUUGACUAUAUAACCUUAUGGCCGCAGCUUGGUCAGACAUUCAGUGUUCAGCUUCUGCUUCAGACAGCACCG--AGUUA-UCCUCCAAUCAA .......(((((..(((((((((.(((((((.(((((......)))))...))))))).)..))))))........(((....)))...))..)))))(--((...-..)))....... ( -28.30) >DroPse_CAF1 42350 118 - 1 CCAAUUAGGUGUCAUCAAUGUCAUUGGAUCGUGUAUAAAAUCCAAUGGCCCGAGACGGGUCAGACAUUCAGUGUUUGGCUUUAGCCCAAGACAGCAACUGCAGUAAAUCCAACUAA-AA ....((((.((((...(((((((((((((..........)))))))((((((...)))))).))))))........(((....)))...))))((....))...........))))-.. ( -31.70) >DroEre_CAF1 46308 104 - 1 CCAAUUAGGUGUCAUCAAUGUCACCAAGUAGCGUAUAAAAUC-CACGGCCCACACUGGGUCAGACAUUCAGUGUUUGGCUUCGGCUCAAGACAGCA------GUUAAUC--------AA .......((((.((....)).)))).....((..........-...(((((.....))))).........((.((.(((....))).)).)).)).------.......--------.. ( -26.80) >DroYak_CAF1 47399 104 - 1 CCAAUUAGGUGUCAUCAAUGUCACCAAGUAGCGUAUAAAAUC-AACGGCUCACACCGGAUCAGACAUUCAGUGUUUGACUUCGGCUCAAGACAGCA------GUUAAUU--------AA ..((((((.(((......((((.....(((((((........-.)).))).)).(((((((((((((...)))))))).))))).....)))))))------.))))))--------.. ( -23.50) >DroPer_CAF1 42446 118 - 1 CCAAUUAGGUGUCAUCAAUGUCAUUGGAUCGCGUAUAAAAUCCAAUGGCCCGAGACGGGUCAGACAUUCAGUGUUUGGCUUCAGCCCAAGACAGCAACUGCAGUAAAUCCAACUAA-AA ....((((.((((...(((((((((((((..........)))))))((((((...)))))).))))))........(((....)))...))))((....))...........))))-.. ( -31.70) >consensus CCAAUUAGGUGUCAUCAAUGUCACCAAGUAGCGUAUAAAAUC_AACGGCCCAAAACGGGUCAGACAUUCAGUGUUUGGCUUCGGCUCAAGACAGCA______GUUAAUC________AA .......(.((((...((((((((........))............(((((.....))))).))))))........(((....)))...)))).)........................ (-20.93 = -20.05 + -0.88)

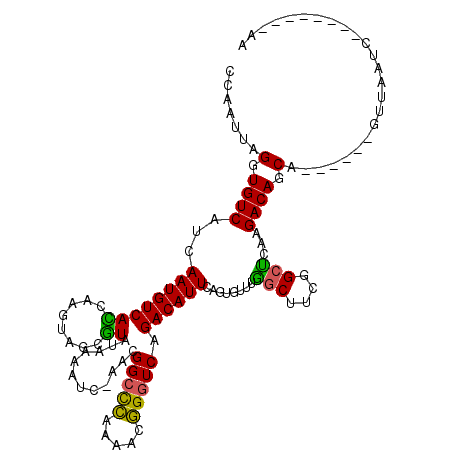
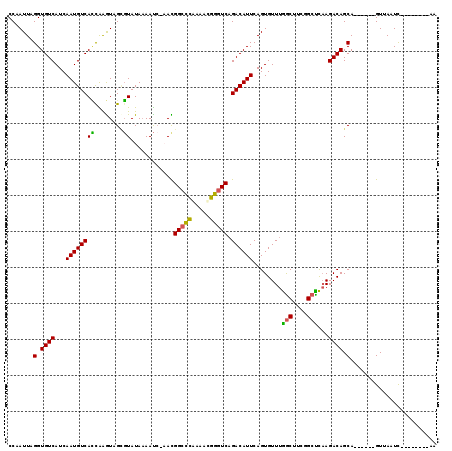
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:49:56 2006