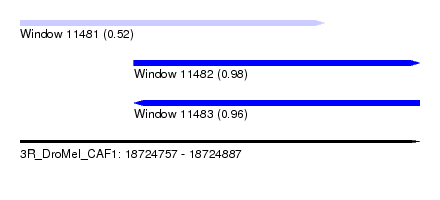
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,724,757 – 18,724,887 |
| Length | 130 |
| Max. P | 0.984934 |

| Location | 18,724,757 – 18,724,856 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 85.31 |
| Mean single sequence MFE | -36.32 |
| Consensus MFE | -26.20 |
| Energy contribution | -27.60 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.72 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.521704 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18724757 99 + 27905053 GCCAA---AGCCAGGGCAGCGCAGCCUAGUGUCUAACAACUUGAGU---UGGUAAUGCCCAUUUCGCCGUGGCACAGUUGUCCUGAGGCGAACUGGGAGAGAG-CC (((((---..(.(((((......)))..((.....))..)).)..)---))))....((((.((((((..((.((....))))...)))))).))))......-.. ( -32.10) >DroSec_CAF1 158432 102 + 1 GCCAA---AGCCAGGGCAGCGCAGCCUAGUGUCUAACAACUUGAGUUGUUGGUAAUGCCCAAUUCGCCGUGGCACAGUUGUCCUGCGGCGAACUGGGAGAGAG-CC ((...---..((.(((((((((......))))((((((((....))))))))...)))))..(((((((..(.((....)).)..)))))))...)).....)-). ( -40.60) >DroSim_CAF1 150080 102 + 1 GCCAA---AGCCAGGGCAGCGCAGCCUAGUGUCUAACAACUUGAGUUGUUGGUAAUGCCCAAUUCGCCGUGGCACAGUUGUCCUGCGGCGAACUGGGAGAGAG-CC ((...---..((.(((((((((......))))((((((((....))))))))...)))))..(((((((..(.((....)).)..)))))))...)).....)-). ( -40.60) >DroEre_CAF1 156181 101 + 1 GCCAAGCAGGGCAGAGCAGAGCAGCCAAG--UCUAACAACUCGAGU---UGGUAAUGCCCAAUUCGCCGUGGCACAGUUGUCCUGCGGCGAACUGGGAGAGAGUCC ........((((...........(((((.--((.........)).)---))))....((((.(((((((..(.((....)).)..))))))).)))).....)))) ( -36.70) >DroYak_CAF1 156877 81 + 1 GCCAA-----------------AGCCA-----CCAACAACUUGAGU---UGGUAAUGCCCAAUUCGCCGUGGCACAGUUGUCCUGCGGCGAACUGGGAGAGAGACC .....-----------------....(-----(((((.......))---))))....((((.(((((((..(.((....)).)..))))))).))))......... ( -31.60) >consensus GCCAA___AGCCAGGGCAGCGCAGCCUAGUGUCUAACAACUUGAGU___UGGUAAUGCCCAAUUCGCCGUGGCACAGUUGUCCUGCGGCGAACUGGGAGAGAG_CC ((((.........((((......))))...........((....))...))))....((((.((((((((((.((....)).)))))))))).))))......... (-26.20 = -27.60 + 1.40)


| Location | 18,724,794 – 18,724,887 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 77.28 |
| Mean single sequence MFE | -37.87 |
| Consensus MFE | -22.77 |
| Energy contribution | -24.15 |
| Covariance contribution | 1.38 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.99 |
| SVM RNA-class probability | 0.984934 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18724794 93 + 27905053 UUGAGU---UGGUAAUGCCCAUUUCGCCGUGGCACAGUUGUCCUGAGGCGAACUGGGAGAGAG-CCAGCCA------UCUUUCGCUCUCUGAGCAGUGCCCUC--- ..(((.---.((((.((((((.((((((..((.((....))))...)))))).))).((((((-(.((...------...)).)))))))..))).)))))))--- ( -35.60) >DroSec_CAF1 158469 87 + 1 UUGAGUUGUUGGUAAUGCCCAAUUCGCCGUGGCACAGUUGUCCUGCGGCGAACUGGGAGAGAG-CCAGCCA------CUUUUCG---------CAGUGCCCUC--- ..((((.((((((....((((.(((((((..(.((....)).)..))))))).)))).....)-))))).)------)))....---------..........--- ( -36.30) >DroSim_CAF1 150117 96 + 1 UUGAGUUGUUGGUAAUGCCCAAUUCGCCGUGGCACAGUUGUCCUGCGGCGAACUGGGAGAGAG-CCAGCCA------CUUUUCGCUCUCUGAGCAGUGUCCGC--- ..((((.((((((....((((.(((((((..(.((....)).)..))))))).)))).....)-))))).)------)))...((((...)))).........--- ( -40.50) >DroEre_CAF1 156219 94 + 1 UCGAGU---UGGUAAUGCCCAAUUCGCCGUGGCACAGUUGUCCUGCGGCGAACUGGGAGAGAGUCCAGCCA------CUGUUCGCUCUGUGAGCAGUGCUCUC--- ....((---(((.....((((.(((((((..(.((....)).)..))))))).)))).......)))))((------((((((((...)))))))))).....--- ( -44.00) >DroYak_CAF1 156895 94 + 1 UUGAGU---UGGUAAUGCCCAAUUCGCCGUGGCACAGUUGUCCUGCGGCGAACUGGGAGAGAGACCAGCCA------CUUUGCGCUCUCUGAGCAGCGCCCUC--- .((.((---((((....((((.(((((((..(.((....)).)..))))))).))))......))))))))------....((((((.....).)))))....--- ( -40.80) >DroAna_CAF1 143706 96 + 1 AUUAGU---CCAUAAUUCCUAAUUCGCCGUGGCACACUUGGCUCGCGGCGAAAAGG-------UCCUGCCAGGACAACCAUGCUCUCUCUGUUUGGCCUUCUCUCU ....((---((...........(((((((((((.(....))).)))))))))..((-------.....)).))))..(((.((.......)).))).......... ( -30.00) >consensus UUGAGU___UGGUAAUGCCCAAUUCGCCGUGGCACAGUUGUCCUGCGGCGAACUGGGAGAGAG_CCAGCCA______CUUUUCGCUCUCUGAGCAGUGCCCUC___ .........(((.....((((.((((((((((.((....)).)))))))))).)))).......)))................((((...))))............ (-22.77 = -24.15 + 1.38)


| Location | 18,724,794 – 18,724,887 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 77.28 |
| Mean single sequence MFE | -35.30 |
| Consensus MFE | -20.81 |
| Energy contribution | -22.97 |
| Covariance contribution | 2.15 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.961982 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18724794 93 - 27905053 ---GAGGGCACUGCUCAGAGAGCGAAAGA------UGGCUGG-CUCUCUCCCAGUUCGCCUCAGGACAACUGUGCCACGGCGAAAUGGGCAUUACCA---ACUCAA ---((((((...((.((.....(....).------))))..)-))))).((((.((((((...((((....)).))..)))))).))))........---...... ( -32.20) >DroSec_CAF1 158469 87 - 1 ---GAGGGCACUG---------CGAAAAG------UGGCUGG-CUCUCUCCCAGUUCGCCGCAGGACAACUGUGCCACGGCGAAUUGGGCAUUACCAACAACUCAA ---((((((.(..---------(.....)------..)...)-))))).((((((((((((..(.((....)).)..))))))))))))................. ( -36.50) >DroSim_CAF1 150117 96 - 1 ---GCGGACACUGCUCAGAGAGCGAAAAG------UGGCUGG-CUCUCUCCCAGUUCGCCGCAGGACAACUGUGCCACGGCGAAUUGGGCAUUACCAACAACUCAA ---((((...))))..(((((((.(....------....).)-))))))((((((((((((..(.((....)).)..))))))))))))................. ( -37.10) >DroEre_CAF1 156219 94 - 1 ---GAGAGCACUGCUCACAGAGCGAACAG------UGGCUGGACUCUCUCCCAGUUCGCCGCAGGACAACUGUGCCACGGCGAAUUGGGCAUUACCA---ACUCGA ---(((((..(.((.(((..........)------)))).)..))))).((((((((((((..(.((....)).)..))))))))))))........---...... ( -37.20) >DroYak_CAF1 156895 94 - 1 ---GAGGGCGCUGCUCAGAGAGCGCAAAG------UGGCUGGUCUCUCUCCCAGUUCGCCGCAGGACAACUGUGCCACGGCGAAUUGGGCAUUACCA---ACUCAA ---(((.(((((........)))))....------....((((......((((((((((((..(.((....)).)..))))))))))))....))))---.))).. ( -39.20) >DroAna_CAF1 143706 96 - 1 AGAGAGAAGGCCAAACAGAGAGAGCAUGGUUGUCCUGGCAGGA-------CCUUUUCGCCGCGAGCCAAGUGUGCCACGGCGAAUUAGGAAUUAUGG---ACUAAU ...(((((((((...(((.((.(((...))).)))))...)).-------)))))))((((.(.(((....).))).))))..(((((.........---.))))) ( -29.60) >consensus ___GAGGGCACUGCUCAGAGAGCGAAAAG______UGGCUGG_CUCUCUCCCAGUUCGCCGCAGGACAACUGUGCCACGGCGAAUUGGGCAUUACCA___ACUCAA ...(((.....(((((...))))).........................((((((((((((..(.((....)).)..))))))))))))............))).. (-20.81 = -22.97 + 2.15)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:37:20 2006