| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,713,593 – 18,713,684 |
| Length | 91 |
| Max. P | 0.836470 |

| Location | 18,713,593 – 18,713,684 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 79.55 |
| Mean single sequence MFE | -27.37 |
| Consensus MFE | -18.80 |
| Energy contribution | -19.92 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.836470 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
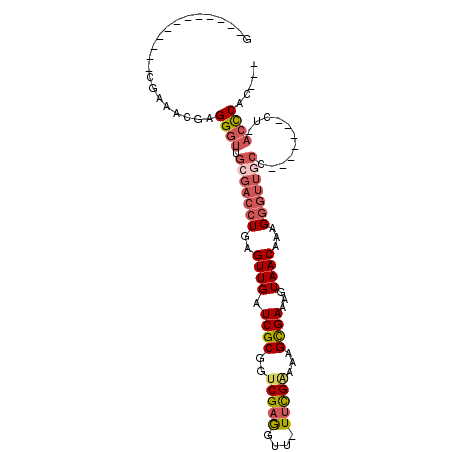
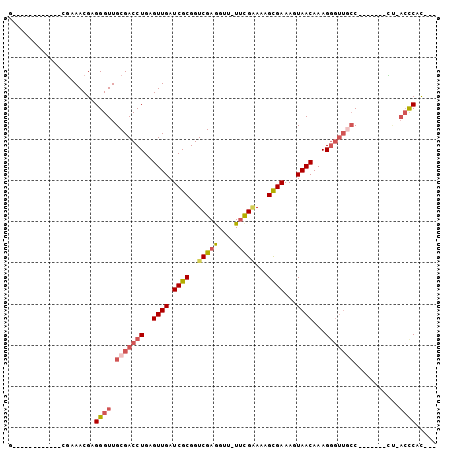
>3R_DroMel_CAF1 18713593 91 + 27905053 G------------CGAAACGAGGGUUGCGACCUGAGUUGAUCGCGGUCGAGGUUUUUCGAAAAGCGAAAGUAACAAAGGGUUGCCAACAACACC-ACCCCUCUC .------------......(((((.((.(.(((..((((.((((..(((((....)))))...))))...))))..)))((((....)))).))-).))))).. ( -29.40) >DroSec_CAF1 147430 76 + 1 G------------CGAAACGAGGGUUGCGACCUGAGUUGAUCGCGGUCGAAGUU-UUCGAAAAGCGAAAGUAACAAAG-----CC-------CUAACCCAC--- .------------........((((((.(..((..((((.((((..((((....-.))))...))))...))))..))-----..-------)))))))..--- ( -21.50) >DroSim_CAF1 139017 80 + 1 G------------CGAAACGAGGGUUGCGACCUGAGUUGAUCGCGGUCGAGGUU-UCCGAAAAGCGAAAGUAACAAAGGGUUGCC-------CU-ACCCAU--- .------------........((((.(((((((..((((.((((..(((.....-..)))...))))...))))...))))))).-------..-))))..--- ( -28.70) >DroEre_CAF1 144835 81 + 1 G------------CGAAACGAGGGUUGCGACCUGAGUUGAUCGCGGUCGAGGUU-UUCGGAAAGCGAAAGUAACAAAGGGUUGCC-------UGCAACCAC--- (------------(....((((..(..((((((((.....))).)))))..)..-))))....)).............(((((..-------..)))))..--- ( -26.70) >DroYak_CAF1 143025 93 + 1 ACGAAAAGCGAAACGAAACGAGGGUUGCGACCUGAGUUGAUCGCGGUCGAGGUU-UUUGGAAAGCGAAAGUAACAAAGGGUUGCC-------CUUACCCAC--- .....................((((.(((((((..((((.((((..((.((...-.)).))..))))...))))...))))))).-------...))))..--- ( -27.10) >DroAna_CAF1 131685 84 + 1 G------------CCAAACGAGGGUUGCGACCUGCGUUGAUCGCGGUCGAGGCU-UUCG-AAAGUGAAAGUAACAAAGGGUUCGCUUCG---GUGUCUCAC--- (------------((...(((((((..(((((.(((.....))))))))..)))-))))-.(((((((..(......)..))))))).)---)).......--- ( -30.80) >consensus G____________CGAAACGAGGGUUGCGACCUGAGUUGAUCGCGGUCGAGGUU_UUCGAAAAGCGAAAGUAACAAAGGGUUGCC_______CU_ACCCAC___ .....................((((.(((((((..((((.((((..(((((....)))))...))))...))))...)))))))...........))))..... (-18.80 = -19.92 + 1.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:37:14 2006