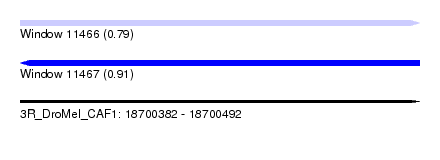
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,700,382 – 18,700,492 |
| Length | 110 |
| Max. P | 0.909182 |

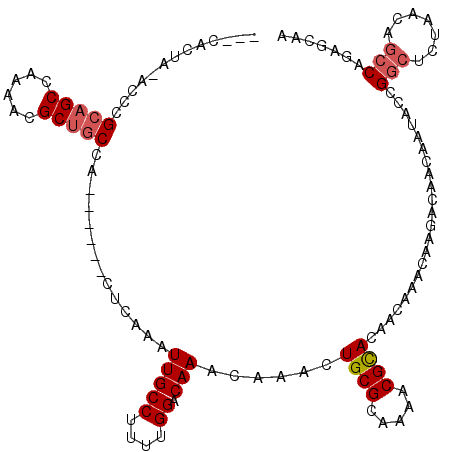
| Location | 18,700,382 – 18,700,492 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 78.45 |
| Mean single sequence MFE | -32.89 |
| Consensus MFE | -21.50 |
| Energy contribution | -21.07 |
| Covariance contribution | -0.43 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.787689 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18700382 110 + 27905053 UUGCCCUGGCUGUUAGAGCCGGUAUUGUUGUCAUGUUUGUUGUGCGUUUUGCGCAGUUUGUUUGUCCAAAAGGCAAUUUGAG------UGGCAGCGUUUUGGCUGCGGGUAUAGUG--- .(((((.((((.....))))(((..(((((((((........(((.(((((.((((.....)))).))))).)))......)------)))))))).....)))..))))).....--- ( -36.64) >DroYak_CAF1 133237 113 + 1 CAGCACUGGUUGUUAGAGUCGGUAUUGUUAUCCUGUUUGUUGUGCGUUUUGCGCAGUUUGUUUGUCCAAAAGGCAAUUUGAG------UGGCAGCGUUUUGGCUGCGGGUGUAGUGUGU ..(((((.((.(((((((.(((((....))))(((((.....(((.(((((.((((.....)))).))))).))).......------.))))).)))))))).)).)))))....... ( -30.12) >DroAna_CAF1 123197 107 + 1 UUGUUUUGGCUGUUAGUGCCGGUAUUAUUGCUUUGUUUGUUGUACGUUUUGCGCAGUUUGUUUGCCCAAAAGGCAAUUUCAGAGAAGUUGGUAGCGUUUUGGCAGCG------------ ........((((((((...((.(((((...(((((...(((((...(((((.((((.....)))).))))).)))))..)))))....))))).))..)))))))).------------ ( -31.90) >consensus UUGCACUGGCUGUUAGAGCCGGUAUUGUUGUCAUGUUUGUUGUGCGUUUUGCGCAGUUUGUUUGUCCAAAAGGCAAUUUGAG______UGGCAGCGUUUUGGCUGCGGGU_UAGUG___ .....(((((.......)))))....................(((.(((((.((((.....)))).))))).)))...............(((((......)))))............. (-21.50 = -21.07 + -0.43)

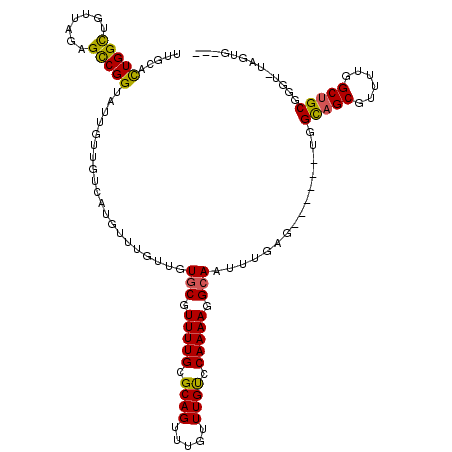
| Location | 18,700,382 – 18,700,492 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 78.45 |
| Mean single sequence MFE | -19.60 |
| Consensus MFE | -11.32 |
| Energy contribution | -12.43 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.909182 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18700382 110 - 27905053 ---CACUAUACCCGCAGCCAAAACGCUGCCA------CUCAAAUUGCCUUUUGGACAAACAAACUGCGCAAAACGCACAACAAACAUGACAACAAUACCGGCUCUAACAGCCAGGGCAA ---.......((((((((......)))))..------.(((..(((((....)).)))......((((.....)))).........)))..........((((.....)))).)))... ( -24.30) >DroYak_CAF1 133237 113 - 1 ACACACUACACCCGCAGCCAAAACGCUGCCA------CUCAAAUUGCCUUUUGGACAAACAAACUGCGCAAAACGCACAACAAACAGGAUAACAAUACCGACUCUAACAACCAGUGCUG .((((((......(((((......)))))..------.........(((((((......)))).((((.....))))........)))........................)))).)) ( -17.50) >DroAna_CAF1 123197 107 - 1 ------------CGCUGCCAAAACGCUACCAACUUCUCUGAAAUUGCCUUUUGGGCAAACAAACUGCGCAAAACGUACAACAAACAAAGCAAUAAUACCGGCACUAACAGCCAAAACAA ------------.(((((......))..................(((.(((((.(((.......))).))))).)))..........))).........(((.......)))....... ( -17.00) >consensus ___CACUA_ACCCGCAGCCAAAACGCUGCCA______CUCAAAUUGCCUUUUGGACAAACAAACUGCGCAAAACGCACAACAAACAAGACAACAAUACCGGCUCUAACAGCCAGAGCAA .............(((((......)))))..............(((((....)).)))......((((.....))))......................(((.......)))....... (-11.32 = -12.43 + 1.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:37:06 2006