| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,691,018 – 18,691,115 |
| Length | 97 |
| Max. P | 0.994553 |

| Location | 18,691,018 – 18,691,115 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 70.24 |
| Mean single sequence MFE | -28.36 |
| Consensus MFE | -8.08 |
| Energy contribution | -10.05 |
| Covariance contribution | 1.97 |
| Combinations/Pair | 1.14 |
| Mean z-score | -3.12 |
| Structure conservation index | 0.28 |
| SVM decision value | 2.49 |
| SVM RNA-class probability | 0.994553 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18691018 97 + 27905053 ------GAGAG--UGUGUGUGUGUGGUGGUGGUUUAAUUUGCUGUUAAAUUGCGCUAAAAAUCCAAAUGCUACCCCC--AACACACACACACACACAUAUAUACAUA--- ------....(--((((((((((((.(((.(((...(((((...(((........))).....)))))...))).))--).))))))))))))).............--- ( -34.60) >DroVir_CAF1 249122 89 + 1 UUCAG----------UGUGUGAAA-GUG-CGCUUUAAUUUGCUGGUAAAUUGCGCUCAAAAUCCAAAUGCUAUCCAG---ACAAAUACAC------AGCCAUAAAUUGGC ...((----------((..(....-)..-))))...(((((((((......(((.............)))...))))---.)))))....------.((((.....)))) ( -19.32) >DroSec_CAF1 120899 98 + 1 UGCUUUGUGAGCGUGUGUGUGUGU-GUGGUGGUUUAAUUUGCUGUUAAAUUGCGCUAAAAAUCCAAAUGCUACCCCC--AACACUCACAC------ACACAUGCAUA--- ..........(((((((((((((.-((((..(((((((.....)))))))..)((.............)).......--..))).)))))------))))))))...--- ( -36.92) >DroSim_CAF1 119963 100 + 1 UGCUUUGUGAGGGUGUGUGUGUGU-GUGGUGGUUUAAUUUGCUGUUAAAUUGCGCUAAAAAUCCAAAUGCUACCCCCACAACACACACAC------AUGCAUACAUA--- .....((((.(.((((((((((((-((((.(((...(((((...(((........))).....)))))...))).)))).))))))))))------)).).))))..--- ( -36.90) >DroWil_CAF1 118523 90 + 1 CUCUCUCUCUC----UGUGUGUGU-GUGCU-GUUUAAUUUGCUGUUAAAUUGCGCUCAAAAUCCAAAUGCUAACCAAGCGGCACCCACAU------GG----GCAU---- ........(((----((((.((((-((((.-(((((((.....))))))).))))............((((.....)))))))).)))).------))----)...---- ( -22.90) >DroMoj_CAF1 134016 89 + 1 UUCAG----------UGUGUGAAA-GUG-CGCUUUAAUUUGCUGGUAAAUUGCGCUCAAAAUCCAAAUGCUAUCCAG---ACAAAUACAG------AGCCAUAAAUUGGC .((((----------((..(....-)..-))))...(((((((((......(((.............)))...))))---.)))))...)------)((((.....)))) ( -19.52) >consensus UUCUG_GUGAG____UGUGUGUGU_GUGGUGGUUUAAUUUGCUGUUAAAUUGCGCUAAAAAUCCAAAUGCUACCCAC__AACACACACAC______AGACAUACAUA___ ...............(((((((((.(((.(((((((((.....))))))))))))........(....)...........)))))))))..................... ( -8.08 = -10.05 + 1.97)

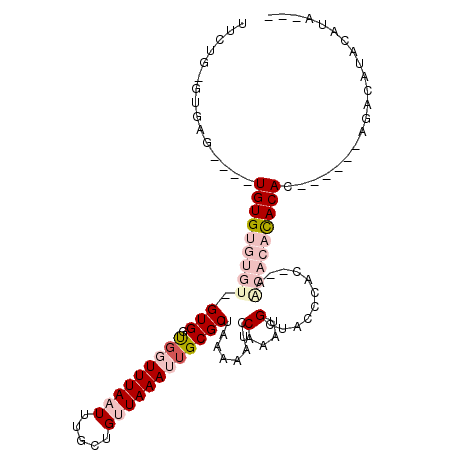

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:37:00 2006