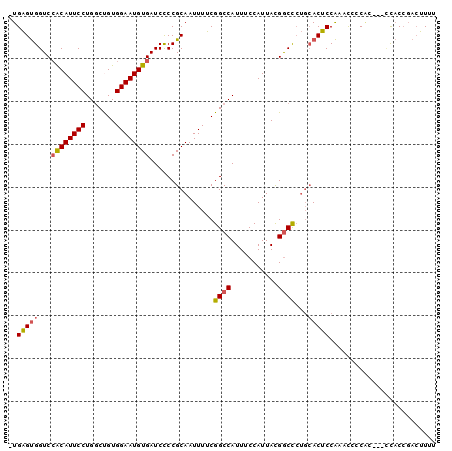
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,674,961 – 18,675,057 |
| Length | 96 |
| Max. P | 0.813995 |

| Location | 18,674,961 – 18,675,057 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 85.89 |
| Mean single sequence MFE | -24.82 |
| Consensus MFE | -21.07 |
| Energy contribution | -21.18 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.813995 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
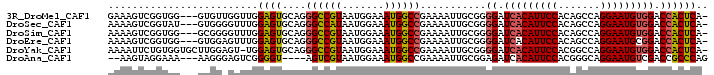
>3R_DroMel_CAF1 18674961 96 + 27905053 -UGAGUGGUCCACAUUCCUGGCUGUGGAAUGUGAUCCCCGCAAUUUUCGGCCAUUUCCAUUACGGCCCUGCACUCCAACCAACAC---CCACCGACUUUC -.((((((..((((((((.......))))))))..))..(((......((((...........)))).)))))))..........---............ ( -24.50) >DroSec_CAF1 104933 96 + 1 -UGAGUGGUCCACAUUCCUGGCUGUGGAAUGUGAUCCCCGCAAUUUUCGGCCAUUUCCAUUAUGGCCCUGCACUCCAAACCCCAC---AUACCGACUUUU -.((((((..((((((((.......))))))))..))..(((......((((((.......)))))).)))))))..........---............ ( -27.70) >DroSim_CAF1 103848 96 + 1 -UGAGUGGUCCACAUUCCUGGCUGUGGAAUGUGAUCCCCGCAAUUUUCGGCCAUUUCCAUUACGGCCCUGCACUCCAAACCCCGC---CCACCGACUUUU -.((((((..((((((((.......))))))))..))..(((......((((...........)))).)))))))..........---............ ( -24.50) >DroEre_CAF1 108057 96 + 1 -UGAGUGGUCCGCAUUCCUGGCUGUGGAAUGUGAUCCCCGCAAUUUUCGGCCAUUUCCAUUACGGCCCUGCACUCCAAACUCCAC---CCACCGACUUUU -.((((((..((((((((.......))))))))..))..(((......((((...........)))).)))))))..........---............ ( -24.10) >DroYak_CAF1 105762 98 + 1 -UGAGUGGUCCACAUUCCUGGCCGUGGAAUGUGAUCCCCGCAAUUUUCGGCCAUUUCCAUUACGGCCCUGCACUCCA-ACUCCAAGCACCACAGAAUUUU -...(((((.((((((((.......))))))))......(((......((((...........)))).)))......-.........)))))........ ( -25.60) >DroAna_CAF1 99633 91 + 1 CUGGGCGGUCGACAUUCCUGCCCGUGGAAUGUGAUCUCCGCAAUUUUCGGCCAUUUCCAUUACGACU----ACCCCGACUCCCUU---UUUCCUACUU-- ..(((..(((((((((((.......))))))).....(((.......)))............)))).----.)))..........---..........-- ( -22.50) >consensus _UGAGUGGUCCACAUUCCUGGCUGUGGAAUGUGAUCCCCGCAAUUUUCGGCCAUUUCCAUUACGGCCCUGCACUCCAAACCCCAC___CCACCGACUUUU ..(((((...((((((((.......))))))))...............((((...........))))...)))))......................... (-21.07 = -21.18 + 0.11)


| Location | 18,674,961 – 18,675,057 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 85.89 |
| Mean single sequence MFE | -30.35 |
| Consensus MFE | -25.27 |
| Energy contribution | -25.25 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.553672 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
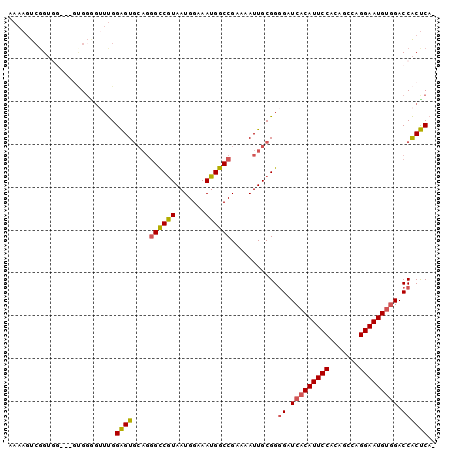
>3R_DroMel_CAF1 18674961 96 - 27905053 GAAAGUCGGUGG---GUGUUGGUUGGAGUGCAGGGCCGUAAUGGAAAUGGCCGAAAAUUGCGGGGAUCACAUUCCACAGCCAGGAAUGUGGACCACUCA- ............---..........((((((((((((((.......)))))).....))))..((.(((((((((.......))))))))).)))))).- ( -30.10) >DroSec_CAF1 104933 96 - 1 AAAAGUCGGUAU---GUGGGGUUUGGAGUGCAGGGCCAUAAUGGAAAUGGCCGAAAAUUGCGGGGAUCACAUUCCACAGCCAGGAAUGUGGACCACUCA- .(((.((.....---...)).))).((((((((((((((.......)))))).....))))..((.(((((((((.......))))))))).)))))).- ( -30.70) >DroSim_CAF1 103848 96 - 1 AAAAGUCGGUGG---GCGGGGUUUGGAGUGCAGGGCCGUAAUGGAAAUGGCCGAAAAUUGCGGGGAUCACAUUCCACAGCCAGGAAUGUGGACCACUCA- .(((.((.(...---.).)).))).((((((((((((((.......)))))).....))))..((.(((((((((.......))))))))).)))))).- ( -32.30) >DroEre_CAF1 108057 96 - 1 AAAAGUCGGUGG---GUGGAGUUUGGAGUGCAGGGCCGUAAUGGAAAUGGCCGAAAAUUGCGGGGAUCACAUUCCACAGCCAGGAAUGCGGACCACUCA- .........(((---((((..((((.(((....((((((.......))))))....))).))))..((.((((((.......)))))).)).)))))))- ( -29.30) >DroYak_CAF1 105762 98 - 1 AAAAUUCUGUGGUGCUUGGAGU-UGGAGUGCAGGGCCGUAAUGGAAAUGGCCGAAAAUUGCGGGGAUCACAUUCCACGGCCAGGAAUGUGGACCACUCA- ..(((((((.......))))))-).((((((((((((((.......)))))).....))))..((.(((((((((.......))))))))).)))))).- ( -32.90) >DroAna_CAF1 99633 91 - 1 --AAGUAGGAAA---AAGGGAGUCGGGGU----AGUCGUAAUGGAAAUGGCCGAAAAUUGCGGAGAUCACAUUCCACGGGCAGGAAUGUCGACCGCCCAG --..........---..(((.((((.(((----..(((((((((......))....)))))))..)))(((((((.......)))))))))))..))).. ( -26.80) >consensus AAAAGUCGGUGG___GUGGGGUUUGGAGUGCAGGGCCGUAAUGGAAAUGGCCGAAAAUUGCGGGGAUCACAUUCCACAGCCAGGAAUGUGGACCACUCA_ .........................((((....((((((.......))))))...........((.(((((((((.......))))))))).)))))).. (-25.27 = -25.25 + -0.02)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:36:43 2006