
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,643,952 – 18,644,067 |
| Length | 115 |
| Max. P | 0.670744 |

| Location | 18,643,952 – 18,644,055 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 79.25 |
| Mean single sequence MFE | -34.68 |
| Consensus MFE | -18.81 |
| Energy contribution | -20.03 |
| Covariance contribution | 1.22 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.670744 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18643952 103 + 27905053 UGACUC------------GGGGUCGAC-GGUCAUCGUUGCAUGCGAAUUUAAUCAAGUCAUGCAAUAGAAUUUACGGUUCCAUUGCCU-UUCGGCCGUCGAUGCCGAUGGUCGUUCC .(((((------------((.((((((-((((.(((((((((((............).)))))))).))......(((......))).-...)))))))))).))))..)))..... ( -41.50) >DroSim_CAF1 70351 103 + 1 UGGCUC------------GGGGUCGAG-GGUCAUCGUUGCAUGCGAAUUUAAUCAAGUCAUGCAAUAGAAUUUACGGUUCCAUUGCCU-UUCGGCCGUCGAUGCCGAUGGUCGUUCG .(((((------------((.(((((.-((((.(((((((((((............).)))))))).))......(((......))).-...)))).))))).))))..)))..... ( -37.30) >DroEre_CAF1 71599 102 + 1 UGGCUC------------GGGGUCGAC-GGUCAUCGUUGCAUGCGAAUUUAAUCAAGUCAUGCAAUAGAAUUUACGGUUCCAUUGCCU-UUCGGCCGUCGAUGCCGAUGCUCGU-CG .(((((------------((.((((((-((((.(((((((((((............).)))))))).))......(((......))).-...)))))))))).)))).)))...-.. ( -42.10) >DroWil_CAF1 71200 90 + 1 ----UU------------GAAGGCGACGGGUCAACGUUGCAUGCGAAUUUAAUCAAGUCAUGCAAUAAAAUUUACGGUUCCUUUGCCUCUUUUGCCUACGAUGCCA----------- ----((------------(.((((((.(((.....(((((((((............).)))))))).........(((......)))))).)))))).))).....----------- ( -22.10) >DroYak_CAF1 72650 114 + 1 UGGGUCGGAGUCGGAGUCGGGGUCGAG-GGUCAUCGUUGCAUGCGAAUUUAAUCAAGUCAUGCAAUAGAAUUUACGGUUCCAUUGCCU-UUCGGCCGUCGAUGCCGAUGCUCGU-CG .........(.(((.(((((.(((((.-((((.(((((((((((............).)))))))).))......(((......))).-...)))).))))).)))))..))).-). ( -40.70) >DroAna_CAF1 70009 95 + 1 -GGUCC------------UGGGUCGCCAGGUCAUCGUUGCAUGCGAAUUUAAUCAAGUCAUGCAAUAGAAUUUACGGUUCCACUGCCC-AUCCACAGAUUUGGCCGAAG-------- -.(.((------------(((....))))).).(((((((((((............).)))))))).)).....(((((...(((...-.....)))....)))))...-------- ( -24.40) >consensus UGGCUC____________GGGGUCGAC_GGUCAUCGUUGCAUGCGAAUUUAAUCAAGUCAUGCAAUAGAAUUUACGGUUCCAUUGCCU_UUCGGCCGUCGAUGCCGAUG_UCGU_CG ..................((.((((...((((...(((((((((............).)))))))).........(((......))).....))))..)))).))............ (-18.81 = -20.03 + 1.22)

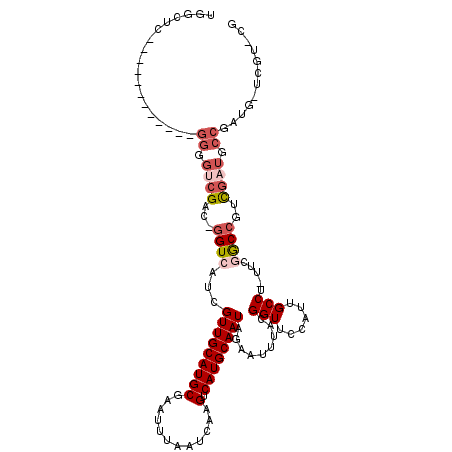

| Location | 18,643,976 – 18,644,067 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.58 |
| Mean single sequence MFE | -23.12 |
| Consensus MFE | -14.98 |
| Energy contribution | -15.07 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.664316 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18643976 91 - 27905053 ACAA--------------ACGAG--------------GACGGAACGACCAUCGGCAUCGACGGCCGAA-AGGCAAUGGAACCGUAAAUUCUAUUGCAUGACUUGAUUAAAUUCGCAUGCA ....--------------..(((--------------.((((..((.....))...((.(..(((...-.)))..).)).))))...)))...((((((....((......)).)))))) ( -24.10) >DroSec_CAF1 73346 91 - 1 ACAA--------------GCGAG--------------GACCGAACGACCAUCGGCAUCGACGGCCGAA-AGGCAAUGGAACCGUAAAUUCUAUUGCAUGACUUGAUUAAAUUCGCAUGCA ....--------------((((.--------------(.((((.......))))).))).).(((...-.)))(((((((.......)))))))(((((....((......)).))))). ( -24.70) >DroSim_CAF1 70375 91 - 1 ACAA--------------GCGAG--------------GACCGAACGACCAUCGGCAUCGACGGCCGAA-AGGCAAUGGAACCGUAAAUUCUAUUGCAUGACUUGAUUAAAUUCGCAUGCA ....--------------((((.--------------(.((((.......))))).))).).(((...-.)))(((((((.......)))))))(((((....((......)).))))). ( -24.70) >DroEre_CAF1 71623 100 - 1 ACA----AUAACCGGCAACCGAC--------------AACCG-ACGAGCAUCGGCAUCGACGGCCGAA-AGGCAAUGGAACCGUAAAUUCUAUUGCAUGACUUGAUUAAAUUCGCAUGCA ...----.......(((..(((.--------------.....-.(((((((((((.......))))).-..(((((((((.......))))))))).)).)))).......)))..))). ( -25.64) >DroWil_CAF1 71221 71 - 1 AC-------------------------------------------------UGGCAUCGUAGGCAAAAGAGGCAAAGGAACCGUAAAUUUUAUUGCAUGACUUGAUUAAAUUCGCAUGCA ..-------------------------------------------------..(((((((..((((.((((.....(....).....)))).))))...))..((......))).)))). ( -10.50) >DroYak_CAF1 72686 118 - 1 GCAACCAACAACCAACAAACGACAAAGGACAAGCGACAACCG-ACGAGCAUCGGCAUCGACGGCCGAA-AGGCAAUGGAACCGUAAAUUCUAUUGCAUGACUUGAUUAAAUUCGCAUGCA (((........((.............))....((((......-.(((((((((((.......))))).-..(((((((((.......))))))))).)).)))).......)))).))). ( -29.06) >consensus ACAA______________ACGAG______________GACCG_ACGACCAUCGGCAUCGACGGCCGAA_AGGCAAUGGAACCGUAAAUUCUAUUGCAUGACUUGAUUAAAUUCGCAUGCA ..................................................(((((.......)))))....(((((((((.......)))))))))........................ (-14.98 = -15.07 + 0.09)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:36:29 2006