
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,621,306 – 18,621,403 |
| Length | 97 |
| Max. P | 0.959267 |

| Location | 18,621,306 – 18,621,403 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 79.81 |
| Mean single sequence MFE | -21.27 |
| Consensus MFE | -15.73 |
| Energy contribution | -16.03 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.959267 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18621306 97 + 27905053 AAAAAUGGG-AACACGAAAAAUAAGUCGGUCAAAUAAAUUUUGCUGUCAAGCGGAAAGCAA-AAGUGCAUAAAAUAUGCAC-AUUGCACUGCGAAUGAGG .....((..-..))..........(.(((.((((.....))))))).)..((((...((((-..(((((((...)))))))-.)))).))))........ ( -21.10) >DroVir_CAF1 43222 82 + 1 ----------UA----UCCAAAAACUCGGUCAUAUACAUUUUGCUGUCAACCGAAAAGCAA-AAGUGCAUAAAGUAUGCAC-AUGGAGC--AGAAUGGGG ----------..----((((....(((.((........(((((((.((....))..)))))-))(((((((...)))))))-)).))).--....)))). ( -20.00) >DroPse_CAF1 53057 93 + 1 AAA-A-----AACAUGGCAAAUAAGUCGGUCAAAUAAAUUUUGCUGUCAGGCGAAAAGCAA-AAGUGCAUAAAAUAUGCACCACUGCCCUGAGAAUGAGG ...-.-----..((.((((.....(.(((.((((.....))))))).)..((.....))..-..(((((((...)))))))...)))).))......... ( -20.60) >DroSec_CAF1 50594 99 + 1 AAAAAUGGGCACCACGAAAAAUAAGUCGUUCAAAUUAAUUUUGCUGUCAAGCGGAAAGCAAAAAGUGCAUAAAAUAUGCAC-UUUGCACUGCGAAUGAGG .........................((((((.......(((((((....))))))).(((.((((((((((...)))))))-)))....))))))))).. ( -26.10) >DroAna_CAF1 46136 88 + 1 AA-AA---G-AACACGAA-----ACUCGGCCAAAUAAAUUUUGCUGUCAAGCGAAAAGCAA-AAGUGCAUAAAAUAUGCAC-AUUGUACUGAGAAUGAGG ..-..---.-........-----.(((((.........(((((((....))))))).((((-..(((((((...)))))))-.)))).)))))....... ( -21.10) >DroPer_CAF1 52132 94 + 1 AAAAA-----AACAUGGUAAAUAAGUCGGUCAAAUAAAUUUUGCUGUCAGGCGAAAAGCAA-AAGUGCAUAAAAUAUGCACCACUGCCCUGAGAAUGAGG .....-----..(((...........(((.((((.....)))))))(((((......(((.-..(((((((...)))))))...))))))))..)))... ( -18.70) >consensus AAAAA_____AACACGAAAAAUAAGUCGGUCAAAUAAAUUUUGCUGUCAAGCGAAAAGCAA_AAGUGCAUAAAAUAUGCAC_AUUGCACUGAGAAUGAGG .........................(((.((.......(((((((....))))))).((((...(((((((...)))))))..)))).....)).))).. (-15.73 = -16.03 + 0.31)

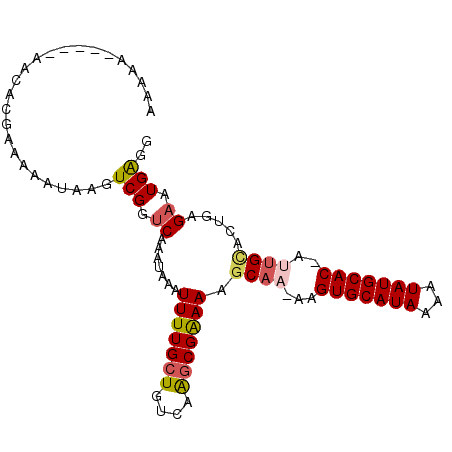
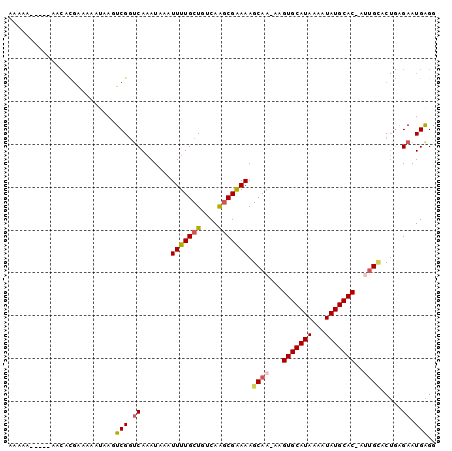
| Location | 18,621,306 – 18,621,403 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 79.81 |
| Mean single sequence MFE | -18.75 |
| Consensus MFE | -10.89 |
| Energy contribution | -10.67 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.707378 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18621306 97 - 27905053 CCUCAUUCGCAGUGCAAU-GUGCAUAUUUUAUGCACUU-UUGCUUUCCGCUUGACAGCAAAAUUUAUUUGACCGACUUAUUUUUCGUGUU-CCCAUUUUU ..(((..((.((.((((.-(((((((...)))))))..-)))).)).))..)))...............((((((........))).)))-......... ( -17.20) >DroVir_CAF1 43222 82 - 1 CCCCAUUCU--GCUCCAU-GUGCAUACUUUAUGCACUU-UUGCUUUUCGGUUGACAGCAAAAUGUAUAUGACCGAGUUUUUGGA----UA---------- ..(((....--((.....-(((((((...)))))))..-..))..(((((((.(((......)))....)))))))....))).----..---------- ( -20.50) >DroPse_CAF1 53057 93 - 1 CCUCAUUCUCAGGGCAGUGGUGCAUAUUUUAUGCACUU-UUGCUUUUCGCCUGACAGCAAAAUUUAUUUGACCGACUUAUUUGCCAUGUU-----U-UUU ........(((((((((.((((((((...)))))))).-))))......)))))..(((((..................)))))......-----.-... ( -19.57) >DroSec_CAF1 50594 99 - 1 CCUCAUUCGCAGUGCAAA-GUGCAUAUUUUAUGCACUUUUUGCUUUCCGCUUGACAGCAAAAUUAAUUUGAACGACUUAUUUUUCGUGGUGCCCAUUUUU ........(((.(((.((-(((((((...)))))))))((((((.((.....)).))))))........................))).)))........ ( -21.30) >DroAna_CAF1 46136 88 - 1 CCUCAUUCUCAGUACAAU-GUGCAUAUUUUAUGCACUU-UUGCUUUUCGCUUGACAGCAAAAUUUAUUUGGCCGAGU-----UUCGUGUU-C---UU-UU ...(((.(((.((.(((.-(((((((...)))))))((-(((((..((....)).))))))).....))))).))).-----...)))..-.---..-.. ( -15.80) >DroPer_CAF1 52132 94 - 1 CCUCAUUCUCAGGGCAGUGGUGCAUAUUUUAUGCACUU-UUGCUUUUCGCCUGACAGCAAAAUUUAUUUGACCGACUUAUUUACCAUGUU-----UUUUU ........(((((((((.((((((((...)))))))).-))))......)))))....................................-----..... ( -18.10) >consensus CCUCAUUCUCAGUGCAAU_GUGCAUAUUUUAUGCACUU_UUGCUUUUCGCUUGACAGCAAAAUUUAUUUGACCGACUUAUUUUUCGUGUU_____UUUUU ...................(((((((...)))))))...(((((.((.....)).)))))........................................ (-10.89 = -10.67 + -0.22)

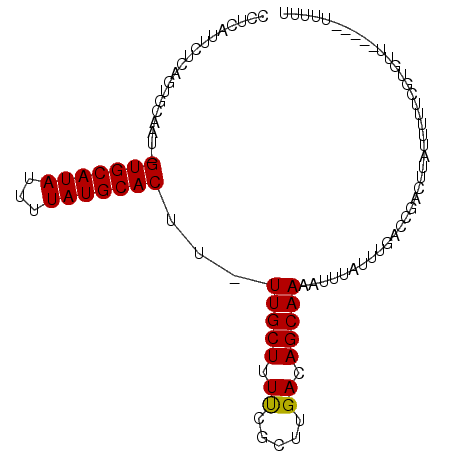
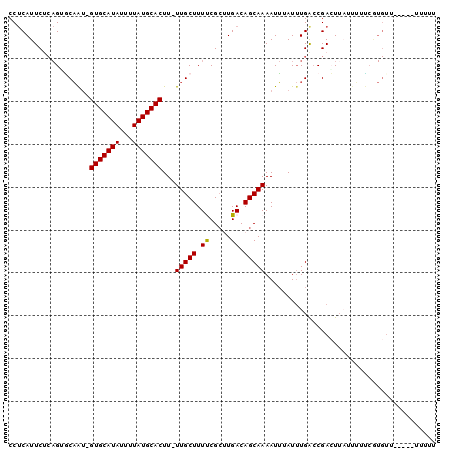
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:36:18 2006