
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,621,133 – 18,621,228 |
| Length | 95 |
| Max. P | 0.995152 |

| Location | 18,621,133 – 18,621,228 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 81.73 |
| Mean single sequence MFE | -25.00 |
| Consensus MFE | -16.12 |
| Energy contribution | -17.36 |
| Covariance contribution | 1.24 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.25 |
| Structure conservation index | 0.64 |
| SVM decision value | 2.55 |
| SVM RNA-class probability | 0.995152 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
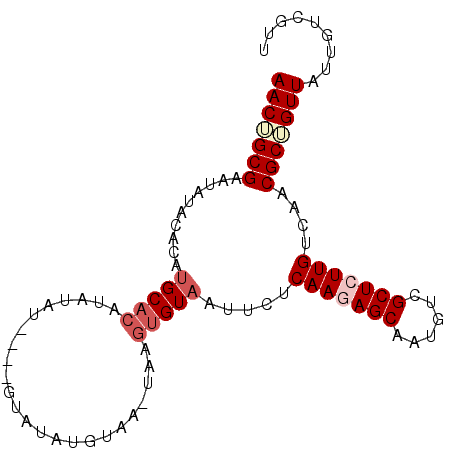
>3R_DroMel_CAF1 18621133 95 + 27905053 AACGGCGAAUAUACAUGUGCACAU-UGUAUAUGUAUAUGUAAGUAAGUGUUAUUCUCAAGAGCAAUGUCGCUCUUGUCAACGCUGUUAUUGUCGUU ((((((((((((((((((((....-.)))))))))))).......((((((.....(((((((......)))))))..))))))....)))))))) ( -35.50) >DroSec_CAF1 50420 96 + 1 AACUGCGAAUAUACACAUGCACAUAUAUGUAUGUAUAUGUAACUAAGUGUAAUUCUCAAGAGCAAUGUCGCUCUUGUCAACGCAGUUAUUGUCGUU ((((((((((.(((((.((.((((((((....))))))))...)).))))))))).(((((((......))))))).....))))))......... ( -30.30) >DroSim_CAF1 47231 92 + 1 AACUGCGAAUAUACACAUGCACAUAUAU----GUAUAUGUAACUAAGUGUAAUUCUCAAGAGCAAUGUCGCUCUUGUCAACGCAGUUAUUGUCGUU ((((((((((.(((((.((.((((((..----..))))))...)).))))))))).(((((((......))))))).....))))))......... ( -27.40) >DroEre_CAF1 47407 79 + 1 AACUGCGA-UACACAUAUGCACAUAUA----------------UAAGUGUAAUUCUCAAGAGCAAUGUCGCUGUUGUCAACGCUGUUAUUGUCUCU (((.(((.-(((((.((((......))----------------)).))))).....(((.(((......))).)))....))).)))......... ( -16.80) >DroYak_CAF1 48357 78 + 1 AACUGCGAAUAUACAAAUGC--GUAUA----------------UAAGUGUAAUUCUCAAGAGCAAUGUCGCUGUUGUCAACGCUGUUAUUGUCUCU (((.(((.((((((......--)))))----------------)............(((.(((......))).)))....))).)))......... ( -15.00) >consensus AACUGCGAAUAUACACAUGCACAUAUAU____GUAUAUGUAA_UAAGUGUAAUUCUCAAGAGCAAUGUCGCUCUUGUCAACGCUGUUAUUGUCGUU (((((((..........(((((........................))))).....(((((((......)))))))....)))))))......... (-16.12 = -17.36 + 1.24)


| Location | 18,621,133 – 18,621,228 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 81.73 |
| Mean single sequence MFE | -21.90 |
| Consensus MFE | -12.82 |
| Energy contribution | -13.90 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.935543 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18621133 95 - 27905053 AACGACAAUAACAGCGUUGACAAGAGCGACAUUGCUCUUGAGAAUAACACUUACUUACAUAUACAUAUACA-AUGUGCACAUGUAUAUUCGCCGUU .........(((.(((....((((((((....))))))))........................(((((((-.((....))))))))).))).))) ( -19.70) >DroSec_CAF1 50420 96 - 1 AACGACAAUAACUGCGUUGACAAGAGCGACAUUGCUCUUGAGAAUUACACUUAGUUACAUAUACAUACAUAUAUGUGCAUGUGUAUAUUCGCAGUU .........((((((.....((((((((....)))))))).(((((((((...((.(((((((......)))))))))..))))).)))))))))) ( -31.60) >DroSim_CAF1 47231 92 - 1 AACGACAAUAACUGCGUUGACAAGAGCGACAUUGCUCUUGAGAAUUACACUUAGUUACAUAUAC----AUAUAUGUGCAUGUGUAUAUUCGCAGUU .........((((((.....((((((((....)))))))).(((((((((...((.((((((..----..))))))))..))))).)))))))))) ( -30.50) >DroEre_CAF1 47407 79 - 1 AGAGACAAUAACAGCGUUGACAACAGCGACAUUGCUCUUGAGAAUUACACUUA----------------UAUAUGUGCAUAUGUGUA-UCGCAGUU ((((.((((.....(((((....)))))..))))))))((.((..(((((.((----------------(........))).)))))-)).))... ( -14.80) >DroYak_CAF1 48357 78 - 1 AGAGACAAUAACAGCGUUGACAACAGCGACAUUGCUCUUGAGAAUUACACUUA----------------UAUAC--GCAUUUGUAUAUUCGCAGUU ((((.((((.....(((((....)))))..))))))))..........(((.(----------------(((((--(....)))))))....))). ( -12.90) >consensus AACGACAAUAACAGCGUUGACAAGAGCGACAUUGCUCUUGAGAAUUACACUUA_UUACAUAUAC____AUAUAUGUGCAUAUGUAUAUUCGCAGUU .........(((.(((....((((((((....))))))))................................((((((....)))))).))).))) (-12.82 = -13.90 + 1.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:36:17 2006