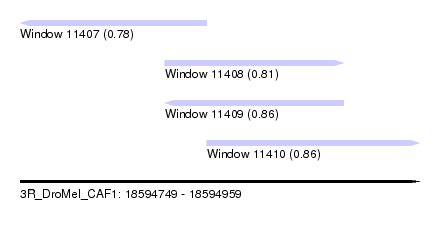
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,594,749 – 18,594,959 |
| Length | 210 |
| Max. P | 0.863145 |

| Location | 18,594,749 – 18,594,847 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 78.77 |
| Mean single sequence MFE | -17.80 |
| Consensus MFE | -10.14 |
| Energy contribution | -11.18 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.783265 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18594749 98 - 27905053 CAAUCCA---------CAGAAACUUUUACCCCAACCAACUUUGAAGCCAUCUUCCACUUUAACCACAACCGAAG---UCAGCUUGGAAAGGGAAGAUCAAAUUCAGGAUG ..((((.---------.(((....)))..............((((...(((((((.((((........((.(((---....))))))))))))))))....)))))))). ( -16.20) >DroSec_CAF1 24318 98 - 1 CAAUCCA---------CAGAAACUUCUACCCCAACCAACUUUGAAGCCAUCUUCCACUUCAACCACAACCGAAG---UCCGCUUGGAAACGGAAGAUCAAAUUCAGGAUG ..((((.---------.(((....)))..............((((...((((((((((((..........))))---)......(....))))))))....)))))))). ( -20.80) >DroSim_CAF1 20758 98 - 1 CAAUCCU---------CAGAAACUUCUACCCCAACCGACUUUGAAGCCAUCUUCCACUUCAACCACAACCGAAG---UUCGCCUGGAAACGGAAGAUCAAAUUCAGGAAG ...((((---------.(((....))).........((.(((((.....(((((((((((..........))))---)......(....)))))))))))).)))))).. ( -19.90) >DroEre_CAF1 19733 107 - 1 CAAUUCAAUACCAAUUGAAAAACUUGUACCACUACCAACUCUAAGGCCAACUUCCACUUCAACAACUACCGAAG---CUCUCUUGGAAACGGAAGACCAAACCCAGGAUG ...((((((....))))))...((((........((........))....(((((.((((..........))))---.......(....))))))........))))... ( -16.20) >DroYak_CAF1 20040 110 - 1 CAAUACAAUUCCAAUUAAAAAACUCGUAGCACUACCAACUCUGAAGCCAACUUCCACAUCAACAACAACCGGAAACAACCUCUUGGAAACGGAGGACCAAACUCAGGAUG ........((((.............(((....))).......((((....))))................))))....(((((.(....)))))).((.......))... ( -15.90) >consensus CAAUCCA_________CAGAAACUUCUACCCCAACCAACUUUGAAGCCAUCUUCCACUUCAACCACAACCGAAG___UCCGCUUGGAAACGGAAGAUCAAAUUCAGGAUG .......................................((((((...(((((((.((((..........))))..........(....))))))))....))))))... (-10.14 = -11.18 + 1.04)

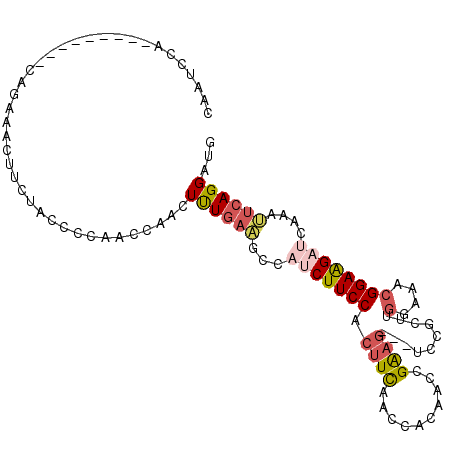
| Location | 18,594,825 – 18,594,919 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 80.46 |
| Mean single sequence MFE | -24.46 |
| Consensus MFE | -10.50 |
| Energy contribution | -12.82 |
| Covariance contribution | 2.32 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.43 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.812785 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18594825 94 + 27905053 GGGUAAAAGUUUCUG---------UGGAUUGGGUACACUUUCAUCAACCCUUUCUUUAAUUAAUUCCAGUGCCCUUGUGGCCAAUGGAAUUAAACCCAUAAUA ((((...........---------.(((..((((............))))..)))....(((((((((..(((.....)))...)))))))))))))...... ( -26.70) >DroSec_CAF1 24394 94 + 1 GGGUAGAAGUUUCUG---------UGGAUUGGGUACACUUUCAUCAACCCUUUCUUUAAUUAAUUCCAGUGCCCUUGUGGCCAAUGGAAUUAAUCCCAUAAUA (((((((....))))---------.(((..((((............))))..)))...((((((((((..(((.....)))...)))))))))))))...... ( -25.20) >DroSim_CAF1 20834 94 + 1 GGGUAGAAGUUUCUG---------AGGAUUGGGUACACUUUCAUCAACCCUUUCUUUAAUUAAUUCCAGUGCCCUUGUGGCCAAUGGAAUUAAUCCCAUAAUA (((..........((---------((((..((((............))))..))))))((((((((((..(((.....)))...)))))))))))))...... ( -28.60) >DroEre_CAF1 19809 103 + 1 UGGUACAAGUUUUUCAAUUGGUAUUGAAUUGGGUACAAUUUCAUCAACCUUUUCUUUAAUUAUUCGCAGUGCCUUUGCAGCCAAUGGAAUUAUUUCUCUAAUA (((...((((..(((.((((((...(((..((((............)))).)))...........((((.....)))).)))))).)))..))))..)))... ( -18.00) >DroYak_CAF1 20119 103 + 1 UGCUACGAGUUUUUUAAUUGGAAUUGUAUUGGGUACAAUAUCAUCCAACCUUUCUUUAAGUAUUCCCACUGCCUUUGGGGCGAAAGGAAUUAAUUCCCUAAUA ......(((((..((..(((((...((((((....))))))..)))))((((((....(((......)))(((.....)))))))))))..)))))....... ( -23.80) >consensus GGGUACAAGUUUCUG_________UGGAUUGGGUACACUUUCAUCAACCCUUUCUUUAAUUAAUUCCAGUGCCCUUGUGGCCAAUGGAAUUAAUCCCAUAAUA (((......................(((..((((............))))..))).....((((((((..(((.....)))...))))))))..)))...... (-10.50 = -12.82 + 2.32)

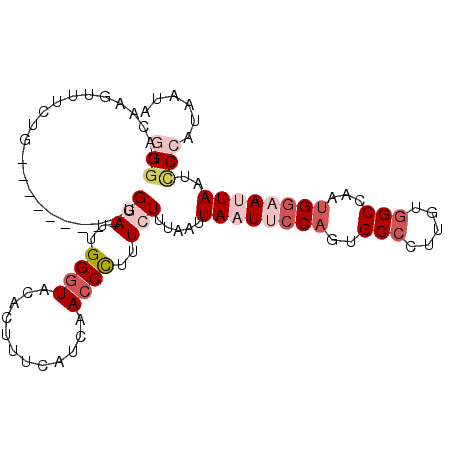
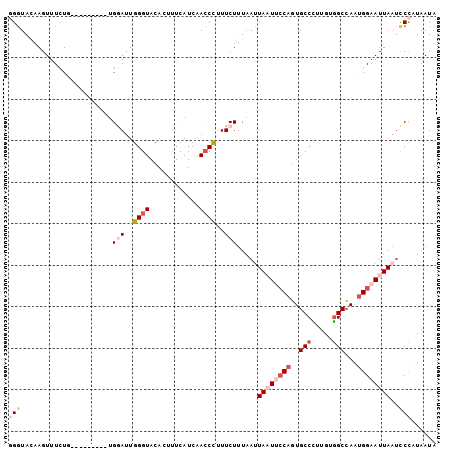
| Location | 18,594,825 – 18,594,919 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 80.46 |
| Mean single sequence MFE | -23.08 |
| Consensus MFE | -13.64 |
| Energy contribution | -15.20 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.858244 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18594825 94 - 27905053 UAUUAUGGGUUUAAUUCCAUUGGCCACAAGGGCACUGGAAUUAAUUAAAGAAAGGGUUGAUGAAAGUGUACCCAAUCCA---------CAGAAACUUUUACCC .....((((((((((((((.((.((....)).)).))))))))..........((((..((....))..))))))))))---------............... ( -26.00) >DroSec_CAF1 24394 94 - 1 UAUUAUGGGAUUAAUUCCAUUGGCCACAAGGGCACUGGAAUUAAUUAAAGAAAGGGUUGAUGAAAGUGUACCCAAUCCA---------CAGAAACUUCUACCC .....((((((((((((((.((.((....)).)).))))))))))).......((((..((....))..))))...)))---------............... ( -27.10) >DroSim_CAF1 20834 94 - 1 UAUUAUGGGAUUAAUUCCAUUGGCCACAAGGGCACUGGAAUUAAUUAAAGAAAGGGUUGAUGAAAGUGUACCCAAUCCU---------CAGAAACUUCUACCC ......(((((((((((((.((.((....)).)).))))))))))...((((.((((..((....))..))))..((..---------..))...)))).))) ( -26.40) >DroEre_CAF1 19809 103 - 1 UAUUAGAGAAAUAAUUCCAUUGGCUGCAAAGGCACUGCGAAUAAUUAAAGAAAAGGUUGAUGAAAUUGUACCCAAUUCAAUACCAAUUGAAAAACUUGUACCA .....(((..((((((.(((..(((((....)).((............))....)))..))).))))))......((((((....))))))...)))...... ( -14.40) >DroYak_CAF1 20119 103 - 1 UAUUAGGGAAUUAAUUCCUUUCGCCCCAAAGGCAGUGGGAAUACUUAAAGAAAGGUUGGAUGAUAUUGUACCCAAUACAAUUCCAAUUAAAAAACUCGUAGCA .....(((.....((((((...(((.....)))...))))))...........(((((((((.(((((....)))))))..)))))))......)))...... ( -21.50) >consensus UAUUAUGGGAUUAAUUCCAUUGGCCACAAGGGCACUGGAAUUAAUUAAAGAAAGGGUUGAUGAAAGUGUACCCAAUCCA_________CAGAAACUUCUACCC ........(((((((((((...(((.....)))..))))))))))).......((((..((....))..)))).............................. (-13.64 = -15.20 + 1.56)


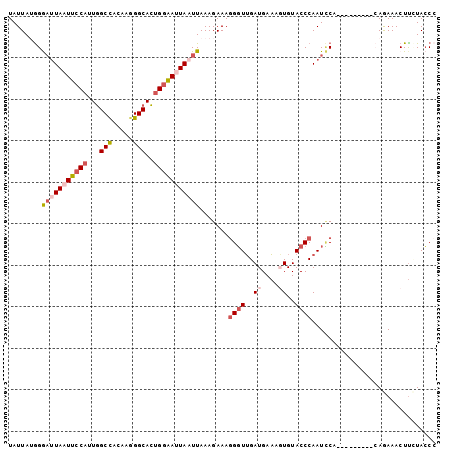
| Location | 18,594,847 – 18,594,959 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 87.95 |
| Mean single sequence MFE | -24.41 |
| Consensus MFE | -18.42 |
| Energy contribution | -18.02 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.863145 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18594847 112 + 27905053 GGUACACUUUCAUCAACCCUUUCUUUAAUUAAUUCCAGUGCCCUUGUGGCCAAUGGAAUUAAACCCAUAAUACCGCUGUGAGGUACAUUCCUUUGACCAUAGUCACUGCCAA ((((.(((....((((............(((((((((..(((.....)))...)))))))))(((((((.......)))).)))........))))....)))...)))).. ( -24.80) >DroSec_CAF1 24416 112 + 1 GGUACACUUUCAUCAACCCUUUCUUUAAUUAAUUCCAGUGCCCUUGUGGCCAAUGGAAUUAAUCCCAUAAUACCGCUGUGAGGUACAUUCCUUUGGCCAUCGUCACUGCCAA (((............))).................(((((.(...((((((((.(((((....((((((.......)))).))...))))).)))))))).).))))).... ( -27.80) >DroSim_CAF1 20856 112 + 1 GGUACACUUUCAUCAACCCUUUCUUUAAUUAAUUCCAGUGCCCUUGUGGCCAAUGGAAUUAAUCCCAUAAUACCGCUGUGAGGUACAUUCCUUUGGCCAUCGUCACUGCCAA (((............))).................(((((.(...((((((((.(((((....((((((.......)))).))...))))).)))))))).).))))).... ( -27.80) >DroEre_CAF1 19840 112 + 1 GGUACAAUUUCAUCAACCUUUUCUUUAAUUAUUCGCAGUGCCUUUGCAGCCAAUGGAAUUAUUUCUCUAAUACCGCUGUGAGGUACACUCCUUUGAUCAUCUUCACUGCUAG (((............)))................((((((((.(..((((....((.((((......)))).))))))..)))))).......(((......))).)))... ( -21.20) >DroYak_CAF1 20150 112 + 1 GGUACAAUAUCAUCCAACCUUUCUUUAAGUAUUCCCACUGCCUUUGGGGCGAAAGGAAUUAAUUCCCUAAUACCGCUUUGAGGUACAUUACUUUGACCAUCUUCACUGCCAA ((((.............((((((....(((......)))(((.....)))))))))..............))))((..((((((.((......)))))....)))..))... ( -20.43) >consensus GGUACACUUUCAUCAACCCUUUCUUUAAUUAAUUCCAGUGCCCUUGUGGCCAAUGGAAUUAAUCCCAUAAUACCGCUGUGAGGUACAUUCCUUUGACCAUCGUCACUGCCAA ((((.................................(((((.(..((((....((.((((......)))).))))))..)))))).......((((....)))).)))).. (-18.42 = -18.02 + -0.40)

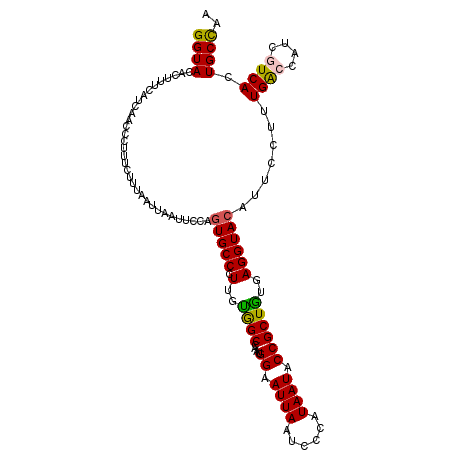
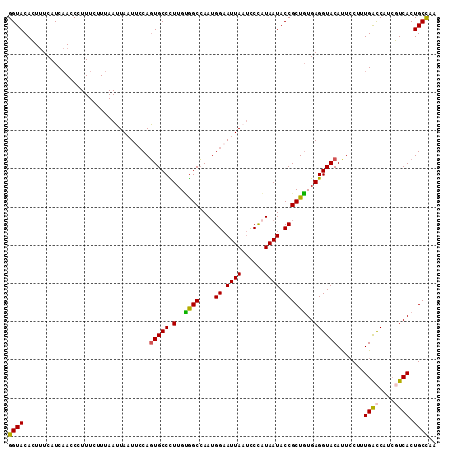
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:36:12 2006