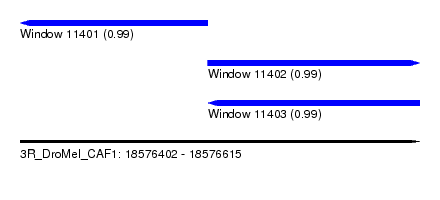
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,576,402 – 18,576,615 |
| Length | 213 |
| Max. P | 0.991045 |

| Location | 18,576,402 – 18,576,502 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 78.62 |
| Mean single sequence MFE | -23.14 |
| Consensus MFE | -17.94 |
| Energy contribution | -18.63 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.99 |
| SVM RNA-class probability | 0.985045 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
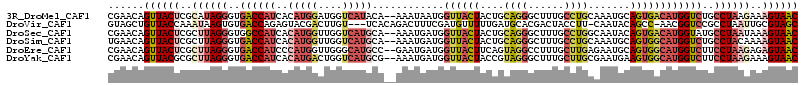
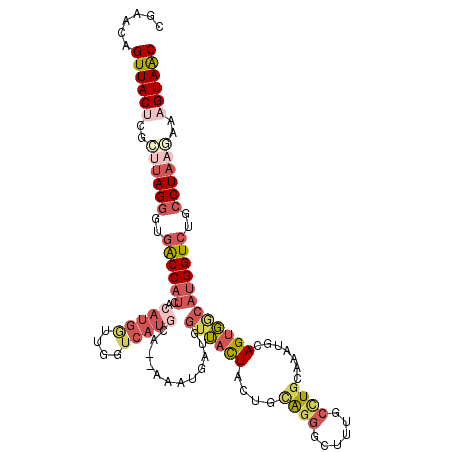
>3R_DroMel_CAF1 18576402 100 - 27905053 AGUUCAAUCACUGCUUUGCUGCCUGGGCUGCCAGGUAACAAAAAGUUAUUAAAAACACAACU--ACUUCUCCUCUUAAGUUAUUUGCAGCUAAAUGGCAGAA ..................(((((..((((((.(((((((...(((.................--.........)))..)))))))))))))....))))).. ( -22.57) >DroSec_CAF1 1446 90 - 1 AGUUCAAUCACUGCUUUGCUGCCAGGGUUGCCACGAAAUGA-AAGUUAUUAAAAACAUUAU-----------UAUUACGUUAUAUGCAGCUAAAUGGCAGAA ..................((((((.((((((.(((.(((((-(((((......))).)).)-----------)))).))).....))))))...)))))).. ( -20.70) >DroSim_CAF1 1453 90 - 1 AGUUCAAUCACUGCUUUGCUGCCAGGGCUGCCAGGUAAUGA-AAGUUAUUAAAAACAAUAU-----------UCUUACGUUAUUUGUAGCUAAAUGGCAGAA ..................((((((.((((((.((((((((.-(((.((((......)))).-----------.))).))))))))))))))...)))))).. ( -27.40) >DroEre_CAF1 1465 97 - 1 AGUUGAAUCACUGCUUUGCUGCCAGGGCUGCCAGGUACUGA-AAGUAAUA----CCACUAAUUAAUUUCUUCACUUAAGUUAUUUGCAGCUAAAUUGCAGUA .........(((((...(((((...((((...((((...((-((.((((.----......)))).))))...)))).))))....)))))......))))). ( -21.90) >consensus AGUUCAAUCACUGCUUUGCUGCCAGGGCUGCCAGGUAAUGA_AAGUUAUUAAAAACACUAU___________UCUUAAGUUAUUUGCAGCUAAAUGGCAGAA ..................((((((.((((((.(((((((.......................................)))))))))))))...)))))).. (-17.94 = -18.63 + 0.69)


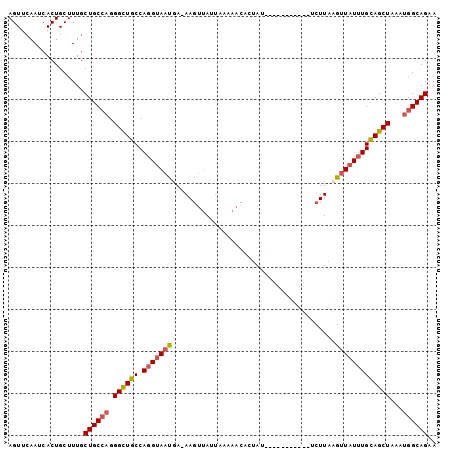
| Location | 18,576,502 – 18,576,615 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 74.13 |
| Mean single sequence MFE | -34.47 |
| Consensus MFE | -18.67 |
| Energy contribution | -20.57 |
| Covariance contribution | 1.89 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.54 |
| SVM decision value | 2.25 |
| SVM RNA-class probability | 0.991045 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18576502 113 + 27905053 GUUACUUUCUUAGGCAGACCAUGUCACUGCAUUUGCAGGCAAAGCCCUGCAGUAGUAACCAUUAUUU--UGUAUGACCAUCCAUGUGAUGGUCACCCUAUGCGAGUAACUGUUCG (((((((.(.((((..(((((.((.(((((...((((((......))))))))))).)).......(--..((((......))))..))))))..)))).).)))))))...... ( -36.60) >DroVir_CAF1 2331 110 + 1 GCUACGCAAUUAGGCGGACCGUU-GGCUGUAUUG-AAGGUAGUCGUGCAUCAAAAACAUCGAAAGUCUGUGA---ACAAGUCGUACUCUGGUCACACUUAUUUGGUAACAGCUAC ....(((......)))......(-((((((((..-((...(((.(((.((((.......(((.....((...---.))..))).....))))))))))..))..)).))))))). ( -25.00) >DroSec_CAF1 1536 113 + 1 GUUACUUUAUUAGGCAUACCAUGUCACUGUAUUGCCAGGCAAAGCCCUGCAGUAGUAACCAUCAUUU--UGCAUGACCAACCAUGUGAUGGCCACCCUAAGCGAGUAACUGUUCG (((((((..(((((...............((((((..(((...)))..))))))((..(((((....--.(((((......))))))))))..)))))))..)))))))...... ( -31.10) >DroSim_CAF1 1543 113 + 1 GUUACUUUUGUAGGCAGACCAUGCCACUGCAUUUGCAGGCAAAGCCCUGCAGUAGUAACCAUCAUUU--UGCAUGACCAACCAUGUGAUGGUCACCCUAAGCGAGUAACUGUUCA (((((((...((((..((((((...(((((...((((((......)))))))))))...........--.(((((......))))).))))))..))))...)))))))...... ( -38.20) >DroEre_CAF1 1562 113 + 1 GUUACUCUCUUAGGAAGACCAUGCCACUGCAUUCUCAAGCAAAGGCCUACUGAAGUAACCAUCAUUC--GGCAUGCCCAACCAUGGGAUGGUCACCCUAAGCGAGUAACUGUUCG (((((((.((((((..((((((.(((.(((........)))..(((...(((((..........)))--))...)))......))).))))))..)))))).)))))))...... ( -43.00) >DroYak_CAF1 1449 113 + 1 GUUACUUUCUUAGGAAGACCAUGCCACUUCAUUCGCAAGCAAAGCCCUACGGUAGUAACCAUCAUUU--CGCAUGACCAGUCAUGUGAUGGUCACCCUAAGCGCGUAACUGUUCG (((((...((((((..((((((((..........))).............(((....)))......(--(((((((....)))))))))))))..))))))...)))))...... ( -32.90) >consensus GUUACUUUCUUAGGCAGACCAUGCCACUGCAUUCGCAGGCAAAGCCCUACAGUAGUAACCAUCAUUU__UGCAUGACCAACCAUGUGAUGGUCACCCUAAGCGAGUAACUGUUCG (((((((.((((((..((((((.....(((........))).............................(((((......))))).))))))..)))))).)))))))...... (-18.67 = -20.57 + 1.89)

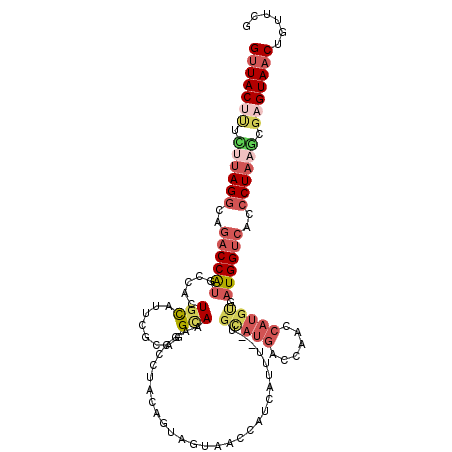
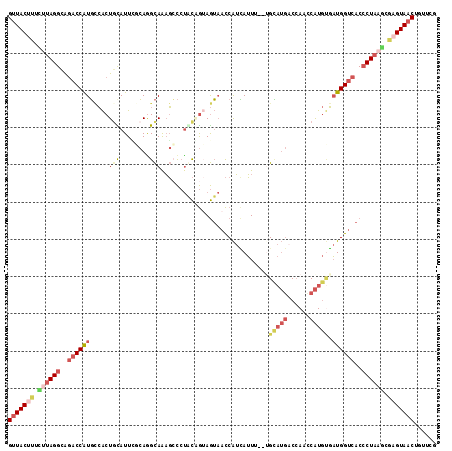
| Location | 18,576,502 – 18,576,615 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 74.13 |
| Mean single sequence MFE | -37.40 |
| Consensus MFE | -19.63 |
| Energy contribution | -21.80 |
| Covariance contribution | 2.17 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.52 |
| SVM decision value | 2.18 |
| SVM RNA-class probability | 0.989820 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18576502 113 - 27905053 CGAACAGUUACUCGCAUAGGGUGACCAUCACAUGGAUGGUCAUACA--AAAUAAUGGUUACUACUGCAGGGCUUUGCCUGCAAAUGCAGUGACAUGGUCUGCCUAAGAAAGUAAC ......((((((..(.(((((((((((((.....)))))))))...--........((((((.(((((((......))))))...).))))))........)))).)..)))))) ( -39.20) >DroVir_CAF1 2331 110 - 1 GUAGCUGUUACCAAAUAAGUGUGACCAGAGUACGACUUGU---UCACAGACUUUCGAUGUUUUUGAUGCACGACUACCUU-CAAUACAGCC-AACGGUCCGCCUAAUUGCGUAGC ......((((((((.((.(((.((((.(((.....)))((---(((.((((.......)))).))).))...........-..........-...))))))).)).))).))))) ( -20.50) >DroSec_CAF1 1536 113 - 1 CGAACAGUUACUCGCUUAGGGUGGCCAUCACAUGGUUGGUCAUGCA--AAAUGAUGGUUACUACUGCAGGGCUUUGCCUGGCAAUACAGUGACAUGGUAUGCCUAAUAAAGUAAC ......((((((.((....((((((((((((((((....)))))..--...)))))))))))...)).((((..((((((.((......)).)).)))).)))).....)))))) ( -35.50) >DroSim_CAF1 1543 113 - 1 UGAACAGUUACUCGCUUAGGGUGACCAUCACAUGGUUGGUCAUGCA--AAAUGAUGGUUACUACUGCAGGGCUUUGCCUGCAAAUGCAGUGGCAUGGUCUGCCUACAAAAGUAAC ......((((((....(((((((((((((((((((....)))))..--...)))))))))))...(((((.((.(((((((....)))..)))).))))))))))....)))))) ( -39.30) >DroEre_CAF1 1562 113 - 1 CGAACAGUUACUCGCUUAGGGUGACCAUCCCAUGGUUGGGCAUGCC--GAAUGAUGGUUACUUCAGUAGGCCUUUGCUUGAGAAUGCAGUGGCAUGGUCUUCCUAAGAGAGUAAC ......(((((((.(((((((.((((((.((((.(((.((....))--.))).))))((((....))))(((.((((........)))).))))))))).))))))).))))))) ( -49.20) >DroYak_CAF1 1449 113 - 1 CGAACAGUUACGCGCUUAGGGUGACCAUCACAUGACUGGUCAUGCG--AAAUGAUGGUUACUACCGUAGGGCUUUGCUUGCGAAUGAAGUGGCAUGGUCUUCCUAAGAAAGUAAC ......(((((...(((((((((((((((((((((....)))))..--...))))))))))......((((((.((((.((.......)))))).))))))))))))...))))) ( -40.70) >consensus CGAACAGUUACUCGCUUAGGGUGACCAUCACAUGGUUGGUCAUGCA__AAAUGAUGGUUACUACUGCAGGGCUUUGCCUGCAAAUGCAGUGGCAUGGUCUGCCUAAGAAAGUAAC ......((((((..((((((..((((((..(((((....)))))............((((((....((((......)))).......))))))))))))..))))))..)))))) (-19.63 = -21.80 + 2.17)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:36:06 2006