| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,563,964 – 18,564,076 |
| Length | 112 |
| Max. P | 0.576620 |

| Location | 18,563,964 – 18,564,076 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 78.54 |
| Mean single sequence MFE | -43.39 |
| Consensus MFE | -23.61 |
| Energy contribution | -23.75 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.576620 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
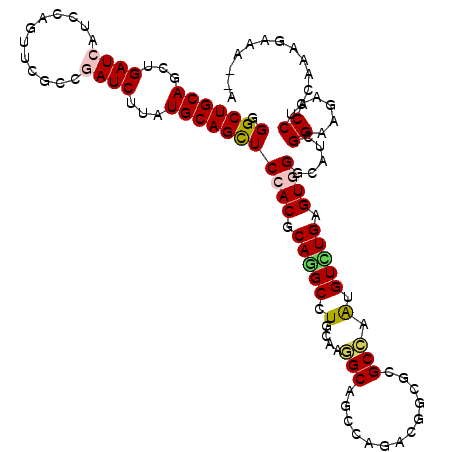
>3R_DroMel_CAF1 18563964 112 + 27905053 AGGCUGCAACUGAUCGUCUAACUCACCGAUCCUGUGCAGUUCCACGCAAGCCUGCAAGGCAGCCAGUCGGCGGGCCAAUGUUUGAGUUGGCAUCGGAAGACCCUAAA----GAAGA .((((((....(((((..........)))))((.(((((.....(....).))))))))))))).(((..(((((((((......)))))).)))...)))......----..... ( -36.10) >DroVir_CAF1 10715 114 + 1 GGGCUGCAGCUGAUCAUCCAGUUCCCCGAUCUUAUGCAGCUCCACACAGGCCUGGAAUGCGGCCAGACGACGUGCCAGUGUUUGCGUGGGCAUUGGAAGACCCUUCAAAUGCA--U .(((((((...((((............))))...)))))))((((.(((((((((.(((((......)).))).)))).))))).))))(((((.((((...)))).))))).--. ( -45.60) >DroPse_CAF1 10504 116 + 1 GGGCUGCAGUUGAUCAUCCAGUUCGCCAAUCUUAUGCAGCUCCACGCAGGCCUGCAGGGCAGCCAGACGUCGCGCCAACGUCUGCGUGGUCAUCGGCAGACCCUGCGAGGAAAUUA ..((((((...(((..............)))...))))))(((.((((((.((((..(((.(((((((((.......))))))).)).)))....))))..)))))).)))..... ( -46.44) >DroYak_CAF1 18032 116 + 1 AGGCUGCAACUGAUCGUCUAACUCGCCGAUCCUGUGCAGCUCAACGCAUGCCUGCAAGGCAGCCAGUCGGCGAGCCAAUGUUUGAGUGGGCAUCGGAAGACCCUAAGAACAAAAGA .(((((((...(((((.(......).)))))...)))))))....(((....)))..(((.(((....)))..)))..(((((....(((..((....)))))...)))))..... ( -40.60) >DroMoj_CAF1 13224 113 + 1 GGGCUGCAGCUGAUCGUCCAGUUCACCUAUCUUAUGCAGCUCCACACAGGCCUGCAAUGCGGCCAAUCGCCGUGCAAGUGUCUGAGUGGGCAUCGGCAGACCCUUUAAUGAGA--- ((((((((((((......)))))...............((.((((.(((((((((...(((((.....))))))).)).))))).))))))....)))).)))..........--- ( -42.10) >DroPer_CAF1 10466 116 + 1 GGGCUGCAGUUGAUCAUCCAGUUCGCCGAUCUUAUGCAGCUCCACGCAGGCCUGCAGGGCAGCCAGACGUCGCGCCAACGUCUGCGUGGUCAUCGGCAGACCCUGCGAGAAAAUAA ((((((((...((((............))))...)))))).)).((((((.((((..(((.(((((((((.......))))))).)).)))....))))..))))))......... ( -49.50) >consensus GGGCUGCAGCUGAUCAUCCAGUUCGCCGAUCUUAUGCAGCUCCACGCAGGCCUGCAAGGCAGCCAGACGGCGCGCCAAUGUCUGAGUGGGCAUCGGAAGACCCUACAAAGAAA__A .(((((((...((((............))))...)))))))((((.(((((.(....(((.............))).).))))).)))).....((.....))............. (-23.61 = -23.75 + 0.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:36:01 2006