
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,543,757 – 18,543,849 |
| Length | 92 |
| Max. P | 0.995859 |

| Location | 18,543,757 – 18,543,849 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 70.63 |
| Mean single sequence MFE | -24.64 |
| Consensus MFE | -5.77 |
| Energy contribution | -6.45 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.23 |
| SVM decision value | 2.62 |
| SVM RNA-class probability | 0.995859 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
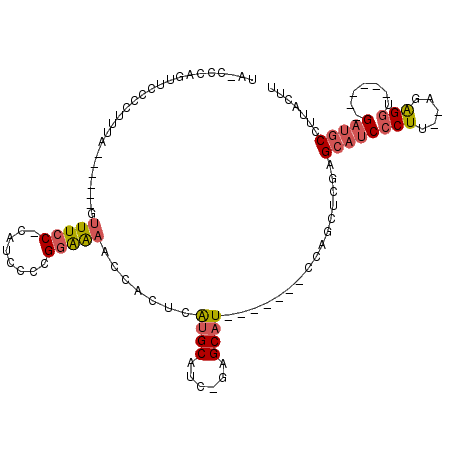
>3R_DroMel_CAF1 18543757 92 + 27905053 UA-CCCAGUUCCCCUUUA------GUUUCC-CAUCCCCGGAGAACCACUCAUGCAUCCGAGCAU-------CCAGCUCGAGCAUCCCUU--AUAGGU------GAUGCCUUACUU ..-...............------(((..(-(......))..)))............(((((..-------...))))).(((((((..--...)).------)))))....... ( -21.00) >DroSec_CAF1 13713 92 + 1 UA-CCCAGUUCCCCUUUA------GUUUCC-CAUCCCCGGAGAAUCACUCAUGCAUCCAAGCAU-------CCAGCUCAAGCAUCCCUU--AUAGGU------GAUGCCUUACUU ..-...((((........------.(((((-.......))))).......((((......))))-------..))))...(((((((..--...)).------)))))....... ( -15.00) >DroEre_CAF1 13497 92 + 1 AA-CACAGUUGC-CUCUA------GAUUCC-CAUCCCCGGAAAUCCACUCAUGCAUC-GAGCAU-------CCAGCUCGAGCAUCCCUUAUAGAGGU------GAUGCCUUACUU ..-....((..(-(((((------((((.(-(......)).)))).....((((.((-((((..-------...))))))))))......)))))).------.))......... ( -30.10) >DroYak_CAF1 13572 97 + 1 UA-CCCAGUUGC-CUCUA------GUUUCC-CAUCCCCGGAAACUCACUCAUGCAUC-GAGCAUCCAGCAUCCAGCUCGAGCAUCCCUU--AGAGGU------GAUGCCUUACUU ..-....((..(-(((((------((((((-.......))))))......((((.((-((((............))))))))))....)--))))).------.))......... ( -31.30) >DroAna_CAF1 13838 101 + 1 GGUUCUAGUCCUCUGUUAGAUCCGGUUUCCACAACUCCGUGAAACCAGUCGAGCG---CAGCAU-------C-ACCGUUAGCAUCCCUU--GCGGCUACAGGAGC-CCCGCAGCU ((.(((((.......))))).))((((((.((......)))))))).....((((---(.....-------.-.......(((.....)--))((((.....)))-)..)).))) ( -25.80) >consensus UA_CCCAGUUCCCCUUUA______GUUUCC_CAUCCCCGGAAAACCACUCAUGCAUC_GAGCAU_______CCAGCUCGAGCAUCCCUU__AGAGGU______GAUGCCUUACUU .........................(((((........))))).......((((......))))................((((((((.....))).......)))))....... ( -5.77 = -6.45 + 0.68)

| Location | 18,543,757 – 18,543,849 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 70.63 |
| Mean single sequence MFE | -31.06 |
| Consensus MFE | -6.53 |
| Energy contribution | -7.45 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.21 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.881462 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
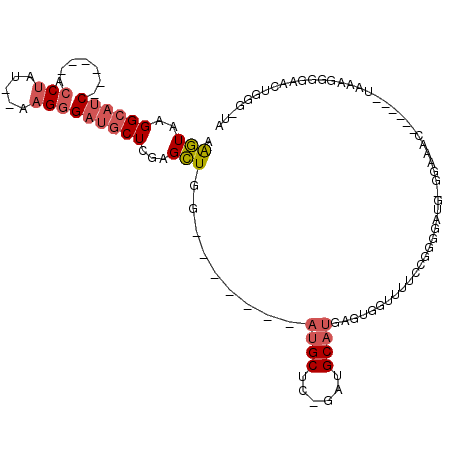
>3R_DroMel_CAF1 18543757 92 - 27905053 AAGUAAGGCAUC------ACCUAU--AAGGGAUGCUCGAGCUGG-------AUGCUCGGAUGCAUGAGUGGUUCUCCGGGGAUG-GGAAAC------UAAAGGGGAACUGGG-UA ..(((.((((((------.((...--..))))))))(((((...-------..)))))..)))......((((((((.....((-(....)------))..))))))))...-.. ( -33.90) >DroSec_CAF1 13713 92 - 1 AAGUAAGGCAUC------ACCUAU--AAGGGAUGCUUGAGCUGG-------AUGCUUGGAUGCAUGAGUGAUUCUCCGGGGAUG-GGAAAC------UAAAGGGGAACUGGG-UA .(((.(((((((------.((...--..)))))))))..)))..-------.((((..(..((....))..((((((.....((-(....)------))..)))))))..))-)) ( -28.00) >DroEre_CAF1 13497 92 - 1 AAGUAAGGCAUC------ACCUCUAUAAGGGAUGCUCGAGCUGG-------AUGCUC-GAUGCAUGAGUGGAUUUCCGGGGAUG-GGAAUC------UAGAG-GCAACUGUG-UU .......(((((------.(((.....))))))))((((((...-------..))))-)).((((...((((((((((....))-))))))------)).((-....)))))-). ( -33.40) >DroYak_CAF1 13572 97 - 1 AAGUAAGGCAUC------ACCUCU--AAGGGAUGCUCGAGCUGGAUGCUGGAUGCUC-GAUGCAUGAGUGAGUUUCCGGGGAUG-GGAAAC------UAGAG-GCAACUGGG-UA ......((((((------.((...--..))))))))..(.(..(.((((..((((..-...)))).....(((((((.......-))))))------)...)-))).)..).-). ( -29.80) >DroAna_CAF1 13838 101 - 1 AGCUGCGGG-GCUCCUGUAGCCGC--AAGGGAUGCUAACGGU-G-------AUGCUG---CGCUCGACUGGUUUCACGGAGUUGUGGAAACCGGAUCUAACAGAGGACUAGAACC ....(((.(-((((((((((((..--..)....))).)))).-)-------).))).---)))..(((((((((((((....))).)))))))).)).................. ( -30.20) >consensus AAGUAAGGCAUC______ACCUAU__AAGGGAUGCUCGAGCUGG_______AUGCUC_GAUGCAUGAGUGGUUUUCCGGGGAUG_GGAAAC______UAAAGGGGAACUGGG_UA .(((..((((((.......(((.....)))))))))...))).........((((......)))).................................................. ( -6.53 = -7.45 + 0.92)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:35:57 2006