
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,542,710 – 18,542,806 |
| Length | 96 |
| Max. P | 0.850658 |

| Location | 18,542,710 – 18,542,806 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 78.69 |
| Mean single sequence MFE | -20.65 |
| Consensus MFE | -17.93 |
| Energy contribution | -17.48 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.14 |
| Mean z-score | -0.92 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.555559 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18542710 96 + 27905053 CGAGUACUGCAGGCCCACCGUGGCGGGCGACAUGGAAAUCACUGAUUCAAUGGGUAAUGCAUCUACAA-AAC---GCA---ACGGAAAA-----------GAGAGAG-AAC--CAA--A ...(((.((((.((((((((((........))))).((((...))))...)))))..))))..)))..-...---...---........-----------.......-...--...--. ( -18.50) >DroVir_CAF1 9500 100 + 1 UGAGUAUUGCAAGCCCACCGUGGCGGGCGACAUGGAAAUCACUGAUUCAAUGGGCAAUGCAUCUACAA--AC---GCA---AAAAAAAAA----------ACAACAGAAAUG-CAAUAA ....(((((((.((((((((((........))))).((((...))))...)))))..(((........--..---)))---.........----------..........))-))))). ( -23.40) >DroGri_CAF1 9915 97 + 1 UGAGUAUUGUAAGCCCACCGUGGCAGGUGACAUGGAAAUCACUGAAUCAAUGGGCAAUGCAUCUACAA--AC---GCA---AAAGGAAAA----------A-AAAG--UACGCCAAUA- ...(((((....(((((...(((..(((((........)))))...))).)))))..(((........--..---)))---.........----------.-..))--))).......- ( -19.70) >DroWil_CAF1 9898 111 + 1 GGAGUAUUGUAAGCCCACUGUGGCGGGCGACAUGGAAAUCACUGAUUCAAUGGGUAAUGCAUCUACAAGAAC---GCAAAAACGGAAACGGAAACAUAACAUAACAA-AAC--CAA--A ((.(((.((((.(((((.((...(((..((........)).)))...)).)))))..))))..)))......---.......((....)).................-..)--)..--. ( -21.10) >DroMoj_CAF1 9629 96 + 1 UGAGUAUUGCAAGCCCACCGUGGCGGGCGACAUGGAAAUCACUGAUUCAAUGGGCAAUGCAUCUACAA--AC---GCA---AAUAAAAUA----------A-AACA--GAUG-CAACA- ...(((((((..((((.(....).))))..(((.(((........))).))).)))))))........--..---(((---.........----------.-....--..))-)....- ( -22.29) >DroAna_CAF1 12728 96 + 1 GGAGUACUGCAGGCCCACCGUGGCGGGCGACAUGGAAAUCACUGAUUCAAUGGGUAAUGCAUCUACAA-AACAAAGCA---ACGGAAAA-------------GAGAG-AAC--CA---A ((.(((.((((.((((((((((........))))).((((...))))...)))))..))))..)))..-.........---.(......-------------)....-..)--).---. ( -18.90) >consensus UGAGUAUUGCAAGCCCACCGUGGCGGGCGACAUGGAAAUCACUGAUUCAAUGGGCAAUGCAUCUACAA__AC___GCA___AAGGAAAAA__________A_AACAG_AAC__CAA__A ...(((.((((.((((((((((........))))).((((...))))...)))))..))))..)))..................................................... (-17.93 = -17.48 + -0.44)

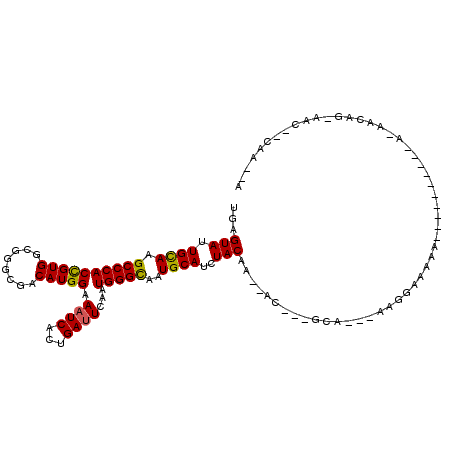
| Location | 18,542,710 – 18,542,806 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 78.69 |
| Mean single sequence MFE | -24.67 |
| Consensus MFE | -17.64 |
| Energy contribution | -17.70 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.850658 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18542710 96 - 27905053 U--UUG--GUU-CUCUCUC-----------UUUUCCGU---UGC---GUU-UUGUAGAUGCAUUACCCAUUGAAUCAGUGAUUUCCAUGUCGCCCGCCACGGUGGGCCUGCAGUACUCG .--.((--(..-.((.((.-----------..(((...---((.---((.-.(((....)))..)).))..)))..)).))...)))(((.((((((....))))))..)))....... ( -22.40) >DroVir_CAF1 9500 100 - 1 UUAUUG-CAUUUCUGUUGU----------UUUUUUUUU---UGC---GU--UUGUAGAUGCAUUGCCCAUUGAAUCAGUGAUUUCCAUGUCGCCCGCCACGGUGGGCUUGCAAUACUCA .(((((-((..........----------.........---(((---((--(....))))))..((((((((.....(((((......)))))......)))))))).))))))).... ( -28.30) >DroGri_CAF1 9915 97 - 1 -UAUUGGCGUA--CUUU-U----------UUUUCCUUU---UGC---GU--UUGUAGAUGCAUUGCCCAUUGAUUCAGUGAUUUCCAUGUCACCUGCCACGGUGGGCUUACAAUACUCA -(((((.....--....-.----------.........---(((---((--(....))))))..((((((((...(((((((......)))).)))...))))))))...))))).... ( -26.00) >DroWil_CAF1 9898 111 - 1 U--UUG--GUU-UUGUUAUGUUAUGUUUCCGUUUCCGUUUUUGC---GUUCUUGUAGAUGCAUUACCCAUUGAAUCAGUGAUUUCCAUGUCGCCCGCCACAGUGGGCUUACAAUACUCC .--..(--((.-((((...(.((((...........((...(((---(((......))))))..)).(((((...))))).....)))).)((((((....))))))..)))).))).. ( -22.20) >DroMoj_CAF1 9629 96 - 1 -UGUUG-CAUC--UGUU-U----------UAUUUUAUU---UGC---GU--UUGUAGAUGCAUUGCCCAUUGAAUCAGUGAUUUCCAUGUCGCCCGCCACGGUGGGCUUGCAAUACUCA -(((((-((..--....-.----------.........---(((---((--(....))))))..((((((((.....(((((......)))))......)))))))).))))))).... ( -28.10) >DroAna_CAF1 12728 96 - 1 U---UG--GUU-CUCUC-------------UUUUCCGU---UGCUUUGUU-UUGUAGAUGCAUUACCCAUUGAAUCAGUGAUUUCCAUGUCGCCCGCCACGGUGGGCCUGCAGUACUCC .---..--...-.....-------------........---......((.-((((((..........(((((...)))))...........((((((....)))))))))))).))... ( -21.00) >consensus U__UUG__GUU_CUGUU_U__________UUUUUCCGU___UGC___GU__UUGUAGAUGCAUUACCCAUUGAAUCAGUGAUUUCCAUGUCGCCCGCCACGGUGGGCUUGCAAUACUCA .....................................................(((..((((...(((((((.....(((((......)))))......)))))))..)))).)))... (-17.64 = -17.70 + 0.06)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:35:55 2006