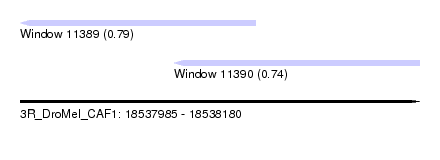
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,537,985 – 18,538,180 |
| Length | 195 |
| Max. P | 0.785128 |

| Location | 18,537,985 – 18,538,100 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.85 |
| Mean single sequence MFE | -33.03 |
| Consensus MFE | -15.55 |
| Energy contribution | -15.17 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.785128 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
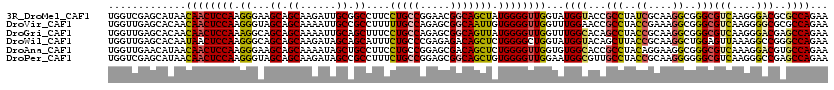
>3R_DroMel_CAF1 18537985 115 - 27905053 ACCGCCUAUCGCAAGGCGGGCGUCAAGGGACGCGCCAGAAAGCAAUUUUACAAAACAAUCAAAAGGGGAAAGGAGACUAUCACGGUAAGU----GCACA-UGCUCAUAUUUCAAUGCAAU .((((((......))))))(((((....)))))(((.((((....))))..................((.((....)).))..)))..(.----((...-.)).)............... ( -29.70) >DroVir_CAF1 4570 113 - 1 ACCGCCUACCGAAAGGCGGGCGUCAAGGGGCGCGCCAGAAAGCAAUUCUACAAAACAAUCAAAAGGGGAAAGGAGACUAUAACGGUAAUUG--U-CAU----CUCCUCACAAAUUAUAAG .((.(((.((....)).(((((((....))))).))((((.....))))..............)))))..((((((..((((......)))--)-..)----)))))............. ( -28.10) >DroPse_CAF1 4648 119 - 1 GUUGCCUACCGCAAGGGGGGCGUCAAGGGCCGAGCCAGAAAGCAAUUUUACAAAACAAUCAAAAGGGGCAAAGAGACCAUCACGGUAGGUCCCUGAACU-GGGUUCAGUUUCAGUUCAAA (.((((..((....))..)))).)..(((((..(((.((((....)))).........((....)))))......(((.....))).))))).((((((-(((......))))))))).. ( -38.30) >DroMoj_CAF1 4631 115 - 1 ACCGCCUAUCGCAAGGCGGGCGUCAAGGGACGCGCCAGAAAGCAAUUUUACAAAACAAUCAAAAGGGGCAAGGAGACUAUAACGGUAACUG--U-UACAAGGCUCCUAUGAGA--AUGAG .((((((......))))))(((((....)))))((......))............((.(((..((((((.((....)).((((((...)))--)-))....)))))).)))..--.)).. ( -36.00) >DroAna_CAF1 8130 100 - 1 ACCGCCUACAGGAAGGCGGGCGUCAAAGGACGUGCCAGAAAGCAAUUUUACAAAACAAUCAAAAGGGGAAAGGAAACUAUAACGGUACGU----------UGCU----------UGCAAU .((((((......))))))..(.(((..((((((((.((((....))))................(..(.((....)).)..))))))))----------)..)----------)))... ( -29.70) >DroPer_CAF1 4632 119 - 1 GUUGCCUACCGCAAGGGGGGCGUCAAGGGCCGAGCCAGAAAGCAAUUUUACAAAACAAUCAAAAGGGGCAAAGAGACCAUCACGGUAGGUCCCUGAAUU-CGGUUCAGUUUCAGUUCAAA (.((((..((....))..)))).)..((((((((...((((....))))..............((((((......(((.....)))..))))))...))-)))))).............. ( -36.40) >consensus ACCGCCUACCGCAAGGCGGGCGUCAAGGGACGCGCCAGAAAGCAAUUUUACAAAACAAUCAAAAGGGGAAAGGAGACUAUAACGGUAAGU___U_AAC__GGCUCCUAUUUCA_UACAAA ((((((..((....))..)))(((...((.....)).((((....)))).........................)))......))).................................. (-15.55 = -15.17 + -0.39)


| Location | 18,538,060 – 18,538,180 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.39 |
| Mean single sequence MFE | -50.80 |
| Consensus MFE | -35.64 |
| Energy contribution | -35.53 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.93 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.744652 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
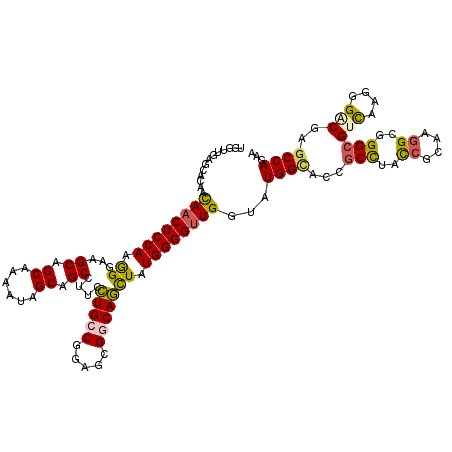
>3R_DroMel_CAF1 18538060 120 - 27905053 UGGUCGAGCAUAACAACUCCAAGGGAAGCAGCAAGAUUGCGGCCUUCCUGCCGGAACGGCAGCUAUGGGGUUGGUAUGGUACCGCCUAUCGCAAGGCGGGCGUCAAGGGACGCGCCAGAA ((((..(.((((.(((((((((((((.((.(((....))).)).)))))((((...)))).....)))))))).)))).).((((((......))))))(((((....)))))))))... ( -53.50) >DroVir_CAF1 4643 120 - 1 UGGUUGAGCACAACAACUCCAAGGGUAGCAGCAAAAUUGCCGCCUUUUUGCCAGAGCGGCAAUUGUGGGGUUGGUUUGGAACCGCCUACCGAAAGGCGGGCGUCAAGGGGCGCGCCAGAA .............((((((((((((..((((.....))))..)))).(((((.....)))))...)))))))).(((((..((((((......))))))(((((....))))).))))). ( -49.40) >DroGri_CAF1 4876 120 - 1 UGGUUGAGCACAACAACUCCAAAGGCAGCAGCAAAAUUGCAGCUUUCCUGCCAGAGCGGCAGUUAUGGGGUUGGUUUGGCACAGCCUACCGCAAGGCGGGCGUCAAGGGACGAGCCAGAA .............(((((((((((((.((((.....)))).))))).(((((.....)))))...)))))))).((((((...(((..((....))..)))(((....)))..)))))). ( -50.10) >DroWil_CAF1 5035 120 - 1 UGGUUGAGCACAAUAACUCCAAGGGCAGCAGCAAGAUAGCAGCAUUUCUGCCCGAGAGACAGCUCUGGGGCUGGUAUGGUACAGCUUACCGCAAGGCUGGAGUUAAAGGCCGGGCCAGAA (((((..((....((((((((.((((....((......))..(((.((.((((.((((....)))).)))).)).))).....)))).((....)).))))))))...))..)))))... ( -45.00) >DroAna_CAF1 8190 120 - 1 UGGUUGAACAUAACAACUCCAAGGGAAGCAGCAAAAUAGCUGCCUUCCUGCCGGAGCGACAGCUCUGGGGUUGGUGUGGCACCGCCUACAGGAAGGCGGGCGUCAAAGGACGUGCCAGAA .............(((((((.(((((.(((((......))))).)))))...(((((....)))))))))))).(.((((.((((((......))))))(((((....))))))))).). ( -56.90) >DroPer_CAF1 4711 120 - 1 UGGUCGAGCAUAACAACUCCAAGGGUAGCAGCAAGAUAGCCGCCUUUCUGCCGGAGCGGCAGCUGUGGGGUUGGAAUGGCGUUGCCUACCGCAAGGGGGGCGUCAAGGGCCGAGCCAGAA ((((((.((....((((((((((((..((.........))..)))).((((((...))))))...))))))))...((((...(((..((....))..)))))))...))))).)))... ( -49.90) >consensus UGGUUGAGCACAACAACUCCAAGGGAAGCAGCAAAAUAGCAGCCUUCCUGCCGGAGCGGCAGCUAUGGGGUUGGUAUGGCACCGCCUACCGCAAGGCGGGCGUCAAGGGACGAGCCAGAA .............((((((((.((...((.((......)).))....(((((.....))))))).))))))))...((((...(((..((....))..)))(((....)))..))))... (-35.64 = -35.53 + -0.11)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:35:53 2006