
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,503,428 – 18,503,539 |
| Length | 111 |
| Max. P | 0.538400 |

| Location | 18,503,428 – 18,503,539 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 78.77 |
| Mean single sequence MFE | -23.43 |
| Consensus MFE | -17.07 |
| Energy contribution | -17.07 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.16 |
| Mean z-score | -0.99 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.538400 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
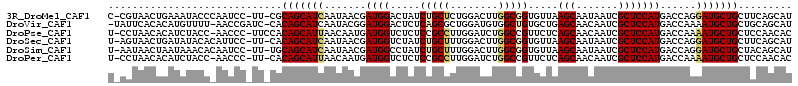
>3R_DroMel_CAF1 18503428 111 + 27905053 C-CGUAACUGAAAUACCCAAUCC-UU-CGCAGCAUCAAUAACGAUGGACUAUCUGCUCUGGACUUGGCGGUGUUAAGCAAUAAUCGCUCCAUGACCAGGAUGCUGCUUCAGCAU .-.....(((((...........-..-.((((((((......((((...))))....((((.(.(((.(((((((.....))).))))))).).)))))))))))))))))... ( -27.53) >DroVir_CAF1 62655 111 + 1 -UAUUCACACAUGUUUU-AACCGAUC-CACAGCAUCAAUACGGAUGGACUCUCAGCGCUGGAUGUGGCUGUGCUGAGCAACAAUCGCUCCAUGACCAAAAUGCUGCUGCAGCAU -........((((....-........-((((((..((...(((.(((....)))...)))..))..))))))..((((.......))))))))......((((((...)))))) ( -25.10) >DroPse_CAF1 42010 111 + 1 U-CCUAACACAUCUACC-AACCC-UUCCACAGCAUUAACAAUGAUGGUCUCUCCGCCUUGGAUCUGGCCGUUCUCAGCAACAAUCGCUCCAUGACCAAAAUGCUGCUCCAACAC .-...............-.....-.....(((((((......(((((((..(((.....)))...)))))))...(((.......)))..........)))))))......... ( -19.60) >DroSec_CAF1 25613 111 + 1 U-AGUAACUGAUAUACACAUUCC-UU-CACAGCAUCAAUAACGAUGGUCUAUCUGCUUUGGACUUGGCGGUGUUAAGCAAUAAUCGCUCCAUGACCAGGAUGCUGCUUCAGCAU .-.....((((............-..-..(((((((........(((((...(((((........))))).....(((.......)))....))))).)))))))..))))... ( -23.17) >DroSim_CAF1 23815 111 + 1 U-AAUAACUAAUAAACACAAUCC-UU-UGCAGCAUCAAUAACGAUGGCCUAUCUGCUUUGGACUUGGCGGUGUUAAGCAAUAAUCGCUCCAUGACCAGGAUGCUGCUACAGCAU .-.....................-..-.((((((((..((((.((.(((..(((.....)))...))).))))))(((.......)))..........))))))))........ ( -25.60) >DroPer_CAF1 42564 110 + 1 U-CCUAACACAUCUACC-AACCC-UU-CACAGCAUUAACAAUGAUGGUCUCUCCGCCUUGGAUCUGGCCGUUCUCAGCAACAAUCGCUCCAUGACCAAAAUGCUGCUCCAACAC .-...............-.....-..-..(((((((......(((((((..(((.....)))...)))))))...(((.......)))..........)))))))......... ( -19.60) >consensus U_ACUAACACAUAUACC_AACCC_UU_CACAGCAUCAAUAACGAUGGUCUAUCUGCUUUGGACUUGGCGGUGCUAAGCAACAAUCGCUCCAUGACCAAAAUGCUGCUCCAGCAU .............................(((((((.......((((.....(((((........))))).....(((.......)))))))......)))))))......... (-17.07 = -17.07 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:35:41 2006