
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,483,236 – 18,483,326 |
| Length | 90 |
| Max. P | 0.980106 |

| Location | 18,483,236 – 18,483,326 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 93.56 |
| Mean single sequence MFE | -26.50 |
| Consensus MFE | -20.34 |
| Energy contribution | -20.78 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.19 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.980106 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
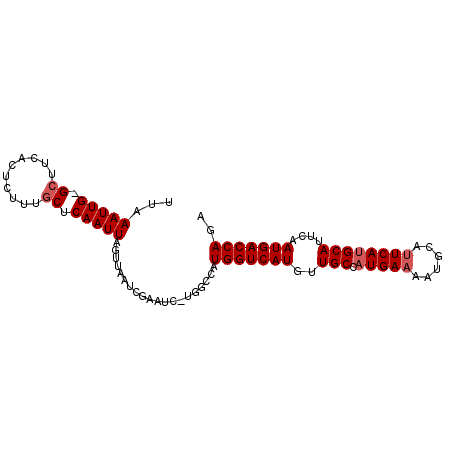
>3R_DroMel_CAF1 18483236 90 + 27905053 UCUGGUCAUUGAAUGCAUGAGUGCAUUUUCAUGGCAACAUGACCAUGGCCA-GAUUCGAUUAACUAAUUGAG--AAGAGUGAAGC-CAAUUUAA (((((((((.(((((((....)))))))(((((....)))))..)))))))-))(((((((....)))))))--...........-........ ( -28.60) >DroSec_CAF1 4912 93 + 1 UCUGGUCAUUGAAUGCAUGAAUGCAUUUUCAUGGCAACAUGACCAUGGCCAGGAUUCGAUUAACUAAUUGAGCAAAGAGUGAAGC-CAAUUUAA (((((((((.(((((((....)))))))(((((....)))))..))))))))).(((((((....))))))).............-........ ( -27.30) >DroSim_CAF1 4895 93 + 1 UCUGGUCAUUGAAUGCAUGAAUGCAUUUUCAUGGCAACAUGACCAUGGCCAGGAUUCGAUUAACUAAUUGAGCAAAGAGUGAAGC-CAAUUUAA (((((((((.(((((((....)))))))(((((....)))))..))))))))).(((((((....))))))).............-........ ( -27.30) >DroEre_CAF1 4793 93 + 1 UCUGGUCAUUGAAUGCCUGAAUACAUUUUCAUGGCAACAUGACCAUGGCCA-GAUUCGAUUAACUAAUUGAGCAAAAUUGGCUGCACAAUUUAA (((((((((.(((((........)))))(((((....)))))..)))))))-))(((((((....)))))))..((((((......)))))).. ( -26.70) >DroYak_CAF1 5078 92 + 1 UCUGGUCAUUGAAUGCAUGAAUACAUUUUCAUGGCAACAUGACCAUGGUCA-GAUUCGAUUAACUAAUUGAGCAAAGAGUGAAGC-CAAUUUAA ..((((((((...(((.(((...(((..(((((....)))))..))).)))-...((((((....)))))))))...))))..))-))...... ( -22.60) >consensus UCUGGUCAUUGAAUGCAUGAAUGCAUUUUCAUGGCAACAUGACCAUGGCCA_GAUUCGAUUAACUAAUUGAGCAAAGAGUGAAGC_CAAUUUAA (((((((((.(((((((....)))))))(((((....)))))..))))))).))(((((((....)))))))...................... (-20.34 = -20.78 + 0.44)



| Location | 18,483,236 – 18,483,326 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 93.56 |
| Mean single sequence MFE | -22.13 |
| Consensus MFE | -17.38 |
| Energy contribution | -17.98 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933734 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18483236 90 - 27905053 UUAAAUUG-GCUUCACUCUU--CUCAAUUAGUUAAUCGAAUC-UGGCCAUGGUCAUGUUGCCAUGAAAAUGCACUCAUGCAUUCAAUGACCAGA ......((-(((....((..--...............))...-.)))))(((((((..(((.((((........)))))))....))))))).. ( -20.43) >DroSec_CAF1 4912 93 - 1 UUAAAUUG-GCUUCACUCUUUGCUCAAUUAGUUAAUCGAAUCCUGGCCAUGGUCAUGUUGCCAUGAAAAUGCAUUCAUGCAUUCAAUGACCAGA ......((-(((....((...(((.....))).....)).....)))))(((((((..(((.(((((......))))))))....))))))).. ( -22.10) >DroSim_CAF1 4895 93 - 1 UUAAAUUG-GCUUCACUCUUUGCUCAAUUAGUUAAUCGAAUCCUGGCCAUGGUCAUGUUGCCAUGAAAAUGCAUUCAUGCAUUCAAUGACCAGA ......((-(((....((...(((.....))).....)).....)))))(((((((..(((.(((((......))))))))....))))))).. ( -22.10) >DroEre_CAF1 4793 93 - 1 UUAAAUUGUGCAGCCAAUUUUGCUCAAUUAGUUAAUCGAAUC-UGGCCAUGGUCAUGUUGCCAUGAAAAUGUAUUCAGGCAUUCAAUGACCAGA ...(((((.((((......)))).))))).............-......(((((((..((((.((((......))))))))....))))))).. ( -25.30) >DroYak_CAF1 5078 92 - 1 UUAAAUUG-GCUUCACUCUUUGCUCAAUUAGUUAAUCGAAUC-UGACCAUGGUCAUGUUGCCAUGAAAAUGUAUUCAUGCAUUCAAUGACCAGA ...(((((-((..........)).))))).((((........-))))..(((((((..(((.(((((......))))))))....))))))).. ( -20.70) >consensus UUAAAUUG_GCUUCACUCUUUGCUCAAUUAGUUAAUCGAAUC_UGGCCAUGGUCAUGUUGCCAUGAAAAUGCAUUCAUGCAUUCAAUGACCAGA ...(((((.((..........)).)))))....................(((((((..(((.(((((......))))))))....))))))).. (-17.38 = -17.98 + 0.60)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:35:35 2006