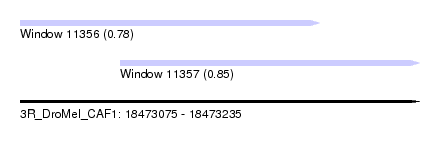
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,473,075 – 18,473,235 |
| Length | 160 |
| Max. P | 0.851283 |

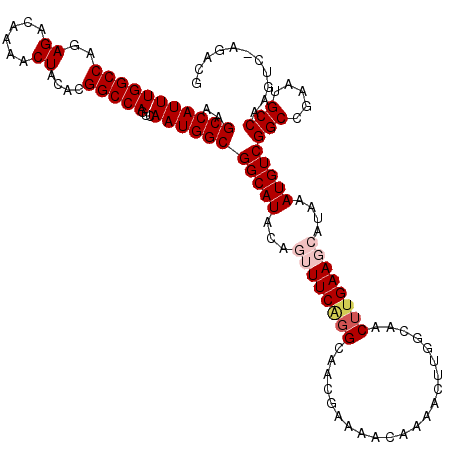
| Location | 18,473,075 – 18,473,195 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.47 |
| Mean single sequence MFE | -38.22 |
| Consensus MFE | -31.57 |
| Energy contribution | -31.49 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.775455 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18473075 120 + 27905053 CUAGGCAGCUUUGCCGGCGGACUGAAUGUCCUGGAGCCGAAAGCCAUUUGGCCAGAGACAAAACUACACGGCCAGUCAAUGGCGGCAUACAGUUUCAGGCAACGAAAACAAAACUUGGCA .......(((.(((((((((((.....))))....)))....(((((((((((..((......))....)))))...))))))))))....((((..(....)..)))).......))). ( -38.90) >DroSec_CAF1 28286 117 + 1 ---GGCUGCUUUGUCGGCGGACUGAAUGUCCUGGAGCCGAAAGCCAUUUGGCCAGAGACAAAACUACACGGCCAGUCAAUGGCGGCAUACAGUUUCGGGCAACGCAAACAAAACUUGGCA ---(..(((.(((((.(.((((((.(((((.((....))...(((((((((((..((......))....)))))...))))))))))).))))))).))))).)))..)........... ( -37.90) >DroSim_CAF1 27996 117 + 1 ---GGCUGCUUUGCCGGCGGACUGAAUGUCCUGGAGCCGAAAGCCAUUUGGCCAGAGACAAAACUACACGGCCAGUCAAUGGCGGCAUACAGUUUCAGGCAACGCAAACAAAACUUGGCA ---(..(((.(((((...((((((.(((((.((....))...(((((((((((..((......))....)))))...))))))))))).))))))..))))).)))..)........... ( -39.60) >DroEre_CAF1 28554 117 + 1 ---GGCAGCUCUGCCAGCGGCCUGAAUGUCCUGGACCCGAAAGCCAUUUGGCCACAGACAAAACUACACGGCCAGUCAAUGGCGGCAUACAAUUUCAGGCAGCGAACACAAGACUUGGCA ---((((....)))).((.(((((((((((.((....))...(((((((((((................)))))...)))))))))))......)))))).))................. ( -36.79) >DroYak_CAF1 29261 117 + 1 ---GCCAGCUUUGCCGGCGGGCUGAAUGUCCUGGACCCGAAAGCCAUUUGGCCAGAGACAAAACUACACGGCCAGUCAAUGGCGGCAUACAAUUUCAGGCAGAGAAAACAGAACUUGGCA ---((((((((((((..((((((.........)).))))...(((((((((((..((......))....)))))...))))))..............)))))))..........))))). ( -37.90) >consensus ___GGCAGCUUUGCCGGCGGACUGAAUGUCCUGGAGCCGAAAGCCAUUUGGCCAGAGACAAAACUACACGGCCAGUCAAUGGCGGCAUACAGUUUCAGGCAACGAAAACAAAACUUGGCA .......((((((((...((((((.(((((.((....))...(((((((((((..((......))....)))))...))))))))))).))))))..)))))..............))). (-31.57 = -31.49 + -0.08)

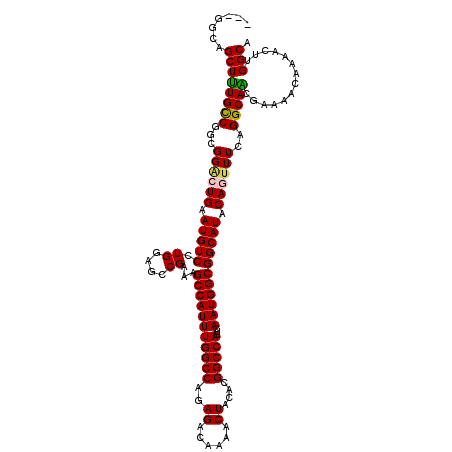
| Location | 18,473,115 – 18,473,235 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.47 |
| Mean single sequence MFE | -32.66 |
| Consensus MFE | -26.91 |
| Energy contribution | -27.99 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.851283 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18473115 120 + 27905053 AAGCCAUUUGGCCAGAGACAAAACUACACGGCCAGUCAAUGGCGGCAUACAGUUUCAGGCAACGAAAACAAAACUUGGCAACUUGAAGCAUAAAUGUCGGCCGAAUGCCAAGUCGAGACG ..(.(((((((((.................((((.....))))(((((...((((((((...(((.........)))....))))))))....)))))))))))))).)........... ( -33.90) >DroSec_CAF1 28323 119 + 1 AAGCCAUUUGGCCAGAGACAAAACUACACGGCCAGUCAAUGGCGGCAUACAGUUUCGGGCAACGCAAACAAAACUUGGCAACUUGAAGCAUAAAUGUCGGCCGACUGCCAAGUC-AGACG ..(((((((((((..((......))....)))))...))))))(((..(((((((((((...(.(((.......))))...)))))))).....)))..)))((((....))))-..... ( -32.60) >DroSim_CAF1 28033 119 + 1 AAGCCAUUUGGCCAGAGACAAAACUACACGGCCAGUCAAUGGCGGCAUACAGUUUCAGGCAACGCAAACAAAACUUGGCAACUUGAAGCAUAAAUGUCGGCCGACUGCCAAGUC-AGACG ..(((((((((((..((......))....)))))...))))))(((..(((((((((((...(.(((.......))))...)))))))).....)))..)))((((....))))-..... ( -33.40) >DroEre_CAF1 28591 119 + 1 AAGCCAUUUGGCCACAGACAAAACUACACGGCCAGUCAAUGGCGGCAUACAAUUUCAGGCAGCGAACACAAGACUUGGCAACACGAACCAUAAAUGUCGGCUGUGUGCCAAGUC-AGACG ..(((((((((((................)))))...))))))............................(((((((((.((((..((.........)).)))))))))))))-..... ( -34.89) >DroYak_CAF1 29298 119 + 1 AAGCCAUUUGGCCAGAGACAAAACUACACGGCCAGUCAAUGGCGGCAUACAAUUUCAGGCAGAGAAAACAGAACUUGGCAACUUGAACCAUAAAUGUCGGCUGAAUGCCAAGUC-AGACG ..(((((((((((..((......))....)))))...))))))(((((.....((((((..(((.........))).....))))))......)))))(((.....))).....-..... ( -28.50) >consensus AAGCCAUUUGGCCAGAGACAAAACUACACGGCCAGUCAAUGGCGGCAUACAGUUUCAGGCAACGAAAACAAAACUUGGCAACUUGAAGCAUAAAUGUCGGCCGAAUGCCAAGUC_AGACG ..(((((((((((..((......))....)))))...))))))(((((...((((((((......................))))))))....)))))(((.....)))........... (-26.91 = -27.99 + 1.08)

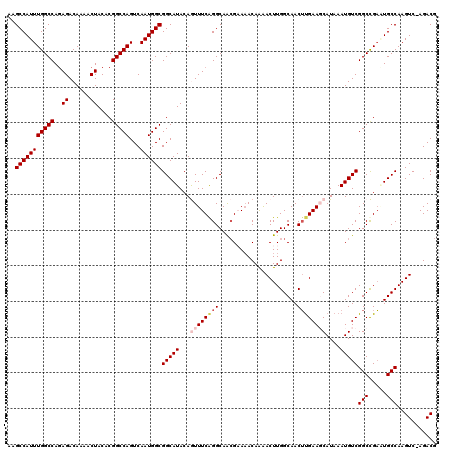
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:35:21 2006