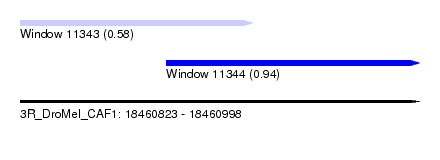
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,460,823 – 18,460,998 |
| Length | 175 |
| Max. P | 0.940165 |

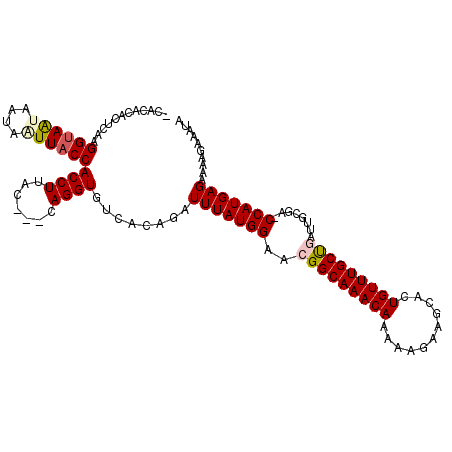
| Location | 18,460,823 – 18,460,925 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 80.67 |
| Mean single sequence MFE | -17.23 |
| Consensus MFE | -12.39 |
| Energy contribution | -12.73 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.576009 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18460823 102 + 27905053 AUGUAUGUAUAAUAUUCUUAAUUUAUGAUAGUAGUU-UUGAGCACUGUCUAUUCAAGCAGAGCAC------------ACACACUCAAGGUAAUAAUAAUUACCACCUUACCACCA .(((.(((.......((..((((.........))))-..))((.((((........)))).))))------------).))).....((((((....))))))............ ( -14.80) >DroSec_CAF1 16198 108 + 1 AU----GUAUAAUGUUCUUAAUUUAUGAUAGUAGACAGUGCCCAAUGUCUAUUCAAACAGAGCACAUGAAUUCAAACACACACUCAAGGUAAUAUUAGUUACCACCUUAC---CA ..----......((((...((((((((...(((((((........)))))))((.....))...))))))))..)))).........((((((....)))))).......---.. ( -18.80) >DroSim_CAF1 15826 96 + 1 AU----GUAUAAUGUUCUUAAUUUAUGAUAGUAGACAGUGCUCAAUGUCUAUUUAAACAGAGCAC------------ACACACUCAAGGUAAUAAUAGUUACCACCUUAC---CA .(----((....((((((...((((....((((((((........)))))))))))).)))))).------------..))).....((((((....)))))).......---.. ( -18.10) >consensus AU____GUAUAAUGUUCUUAAUUUAUGAUAGUAGACAGUGCCCAAUGUCUAUUCAAACAGAGCAC____________ACACACUCAAGGUAAUAAUAGUUACCACCUUAC___CA ............((((((.......(((..(((((((........))))))))))...)))))).......................((((((....))))))............ (-12.39 = -12.73 + 0.34)

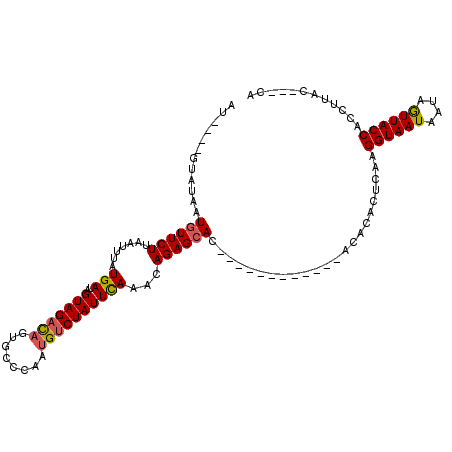
| Location | 18,460,887 – 18,460,998 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 89.09 |
| Mean single sequence MFE | -25.42 |
| Consensus MFE | -19.38 |
| Energy contribution | -19.98 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.940165 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18460887 111 + 27905053 --ACACACUCAAGGUAAUAAUAAUUACCACCUUACCACCAGGUGUCACAGUUUUAUGGAACGGCAAACAAAAAGAAACUUUGUUUGCUGAUUGCGA-CCAUGAGAAAAGAAAUA --.....((((.(((..((((......(((((.......)))))................(((((((((((.......)))))))))))))))..)-)).)))).......... ( -28.00) >DroSec_CAF1 16269 110 + 1 ACACACACUCAAGGUAAUAUUAGUUACCACCUUAC---CAGGUGUCACAGAUUUAUGGAACGGCAAACAAAAAGAAGCACUGUUUGCUGAUUGCGA-CCAUGAGAAAAGAAAUA .......((((.((((((....))))))((((...---.))))(((.((......))...(((((((((...........))))))))).....))-)..)))).......... ( -25.30) >DroSim_CAF1 15887 108 + 1 --ACACACUCAAGGUAAUAAUAGUUACCACCUUAC---CAGGUGUCACAGAUUUAUGGAACGGCAAACAAAAAGAAGCUCUGUUUGCUGAUUGCGA-CCAUGAGAAAAGAAAUA --.....((((.(((..((((.((...(((((...---.)))))..))............(((((((((...........)))))))))))))..)-)).)))).......... ( -26.00) >DroEre_CAF1 16700 106 + 1 -CAAA--CGCAGGGUAAUAAUAUUUACCACCU-AG---CAGGUGUCACAGAUUUAUGGAUGGGCAAACAA-AAGAAGCACUGUUUGCUGAUUGCGAGCCAUGAGAAAAGAAAUA -....--(((..(((((......)))))(((.-..---..)))..............(((.((((((((.-.........)))))))).))))))................... ( -22.40) >DroYak_CAF1 16838 109 + 1 -CAAACACACAAGGCAGCAAUAAUUACCACCUUAC---CAGGUGUCACAGAUUUAUGGAUGGGCAAACAAAAAGAAGCACUGUUUGCCGAUUGCGA-CCAUGAGAAAAGAAAUA -........((.((..(((((......(((((...---.))))).................((((((((...........)))))))).)))))..-)).))............ ( -25.40) >consensus _CACACACUCAAGGUAAUAAUAAUUACCACCUUAC___CAGGUGUCACAGAUUUAUGGAACGGCAAACAAAAAGAAGCACUGUUUGCUGAUUGCGA_CCAUGAGAAAAGAAAUA ............((((((....))))))((((.......))))........(((((((..(((((((((...........)))))))))........))))))).......... (-19.38 = -19.98 + 0.60)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:35:08 2006