
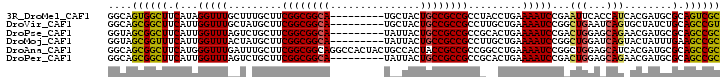
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,446,997 – 18,447,130 |
| Length | 133 |
| Max. P | 0.851306 |

| Location | 18,446,997 – 18,447,090 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 81.25 |
| Mean single sequence MFE | -38.08 |
| Consensus MFE | -23.04 |
| Energy contribution | -22.76 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.742678 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18446997 93 + 27905053 GGCAGUGGCUUCAUAGGUUUGCUUUGCUUCGGCGGCA---------UGCUACUGCCGCCGCCUACCUGAAAAUCCGAAUUCACCAUCACGAUGCGCAGUCGC (((.(((((......(((.((((..((....))))))---------.)))...))))).)))...(((...(((.((........))..)))...))).... ( -27.00) >DroVir_CAF1 8973 93 + 1 GGCAGCGGCUUCAUUGGUUUGCUAUGCUUCGGCGGCA---------UGCUACUGCCGCCGCCUUGCUGAAAAUCCGGCUGAAUCAGUGCUAUCUGCAGCCGU (((.((((...(((((((((.........((((((((---------......))))))))....((((......)))).)))))))))....)))).))).. ( -38.50) >DroPse_CAF1 7140 93 + 1 GGUAGCGGCUUCAUUGGUUUAGUCUGCUUCGGCGGCA---------UAUUACUGCCGCCGCCGCACUGAAAAUCCGACUGGAGCAGAACGAUGCGCAGCCGC ....((((((.(((((........(((..((((((((---------......))))))))..)))(((....(((....))).)))..)))))...)))))) ( -39.30) >DroMoj_CAF1 7018 93 + 1 GGUAGCGGUUUCAUUGGUUUACUAUGCUUCGGCGGCA---------UAUUACUGCCGCCGCCUUGCUGAAAAUCCGGCUGGAUCAGUACUAUUUGAAGCCGC ....(((((((((.((((..(((......((((((((---------......))))))))((..((((......)))).))...)))))))..))))))))) ( -38.90) >DroAna_CAF1 3219 102 + 1 GGCAGCGGCUUCAUGGGUUUGAUUUGCUUCGGCGGCAGGCCACUACUGCCACUACCGCCGCCGGCCUGAAAAUCCGGCUGGAGCAUCACGAUGCGCAGCCGC (((.(((((.(((......)))...((....))(((((.......)))))......)))))..)))........((((((..((((....)))).)))))). ( -44.90) >DroPer_CAF1 9318 93 + 1 GGCAGCGGCUUCAUUGGUUUAGUCUGCUUCGGCGGCA---------UAUUACUGCCGCCGCCGCACUGAAAAUCCGACUGGAGCAGAACGAUGCGCAGCCGC (((.(((((((((((((((((((..((..((((((((---------......))))))))..))))))))...)))).)))))).........))).))).. ( -39.90) >consensus GGCAGCGGCUUCAUUGGUUUACUAUGCUUCGGCGGCA_________UACUACUGCCGCCGCCGCACUGAAAAUCCGACUGGAGCAGAACGAUGCGCAGCCGC ....((((((.(...(((((.........((((((((...............)))))))).........)))))...(((...)))........).)))))) (-23.04 = -22.76 + -0.28)
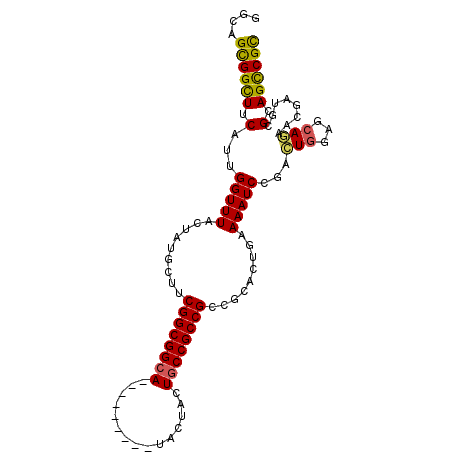

| Location | 18,446,997 – 18,447,090 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 81.25 |
| Mean single sequence MFE | -39.05 |
| Consensus MFE | -22.76 |
| Energy contribution | -23.51 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.851306 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18446997 93 - 27905053 GCGACUGCGCAUCGUGAUGGUGAAUUCGGAUUUUCAGGUAGGCGGCGGCAGUAGCA---------UGCCGCCGAAGCAAAGCAAACCUAUGAAGCCACUGCC (((....)))...((..(((((((........)))..(((((((((((((......---------))))))))..((...))...)))))...))))..)). ( -32.00) >DroVir_CAF1 8973 93 - 1 ACGGCUGCAGAUAGCACUGAUUCAGCCGGAUUUUCAGCAAGGCGGCGGCAGUAGCA---------UGCCGCCGAAGCAUAGCAAACCAAUGAAGCCGCUGCC .(((((((((......)))...))))))......((((..((((((((((......---------)))))))...((...))...........))))))).. ( -36.30) >DroPse_CAF1 7140 93 - 1 GCGGCUGCGCAUCGUUCUGCUCCAGUCGGAUUUUCAGUGCGGCGGCGGCAGUAAUA---------UGCCGCCGAAGCAGACUAAACCAAUGAAGCCGCUACC ((((((...(((.((((((((.(.((((.((.....)).))))(((((((......---------)))))))).))))))....))..))).)))))).... ( -42.10) >DroMoj_CAF1 7018 93 - 1 GCGGCUUCAAAUAGUACUGAUCCAGCCGGAUUUUCAGCAAGGCGGCGGCAGUAAUA---------UGCCGCCGAAGCAUAGUAAACCAAUGAAACCGCUACC ((((.((((.....(((((((((....)))).....((....((((((((......---------))))))))..)).)))))......)))).)))).... ( -35.90) >DroAna_CAF1 3219 102 - 1 GCGGCUGCGCAUCGUGAUGCUCCAGCCGGAUUUUCAGGCCGGCGGCGGUAGUGGCAGUAGUGGCCUGCCGCCGAAGCAAAUCAAACCCAUGAAGCCGCUGCC ((((((...(((..(((((((.(.(((((.........)))))((((((((.(.((....)).)))))))))).))))..))).....))).)))))).... ( -45.90) >DroPer_CAF1 9318 93 - 1 GCGGCUGCGCAUCGUUCUGCUCCAGUCGGAUUUUCAGUGCGGCGGCGGCAGUAAUA---------UGCCGCCGAAGCAGACUAAACCAAUGAAGCCGCUGCC ((((((...(((.((((((((.(.((((.((.....)).))))(((((((......---------)))))))).))))))....))..))).)))))).... ( -42.10) >consensus GCGGCUGCGCAUCGUACUGCUCCAGCCGGAUUUUCAGCAAGGCGGCGGCAGUAACA_________UGCCGCCGAAGCAAAGCAAACCAAUGAAGCCGCUGCC ((((((.((...........(((....)))......((....((((((((...............))))))))..))............)).)))))).... (-22.76 = -23.51 + 0.75)



| Location | 18,447,019 – 18,447,130 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.26 |
| Mean single sequence MFE | -43.93 |
| Consensus MFE | -23.12 |
| Energy contribution | -24.88 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.792024 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18447019 111 + 27905053 UUUGCUUCGGCGGCA---------UGCUACUGCCGCCGCCUACCUGAAAAUCCGAAUUCACCAUCACGAUGCGCAGUCGCUGGCCAUCGGCCAUGGUAACCGGCUGAUUGAGGUCGAUUG .((((((((((((((---------......)))))))(((.....(((........)))(((((..(((((.((........)))))))...)))))....))).....)))).)))... ( -38.40) >DroVir_CAF1 8995 111 + 1 UAUGCUUCGGCGGCA---------UGCUACUGCCGCCGCCUUGCUGAAAAUCCGGCUGAAUCAGUGCUAUCUGCAGCCGUUGGCCAUCUGCCACAGUGACAGGUUGCAUCAAAUCAAGCG ...((((.(((((((---------......)))))))((((..(((......((((((...(((......))))))))).((((.....)))))))..)..)))...........)))). ( -41.20) >DroGri_CAF1 8803 111 + 1 UAUGCUUCGGCGGCA---------UGCUACUGCCGCCGCCUAACUGAAAAUCCGGAUUGAGCAAGGCAAUCUGCAGGCGCAGUCCAUCCGCAACGGUGACGGGCUGCAUUAGAUCCAUUG .((((...(((((((---------......)))))))(((.............((((((.((...((.....))..)).))))))........((....))))).))))........... ( -41.40) >DroWil_CAF1 3747 111 + 1 UGAGCUUCGGCGGCA---------CAAUAUUGCCGCCGCCUGGCUGAAAAUCCGACUGACGCAUUACAAUCUGCAGCCGAUGGCCAUCAGCUAUGGUAACGGGCUGCAUCAGGUCGAGUG (.((((.((((((((---------......))))))))...)))).).....(((((((............(((((((....(((((.....)))))....)))))))))).)))).... ( -43.70) >DroMoj_CAF1 7040 111 + 1 UAUGCUUCGGCGGCA---------UAUUACUGCCGCCGCCUUGCUGAAAAUCCGGCUGGAUCAGUACUAUUUGAAGCCGCUGCCCAUCGGCCACUGUGACGGGCUGCAUCAGAUCAAGCG ...((((((((((((---------......)))))))).....((((......((((...((((......))))))))((.((((.(((.......))).)))).)).))))...)))). ( -42.50) >DroAna_CAF1 3241 120 + 1 UUUGCUUCGGCGGCAGGCCACUACUGCCACUACCGCCGCCGGCCUGAAAAUCCGGCUGGAGCAUCACGAUGCGCAGCCGCUGGCCAUCGGCCAUGGUCACCGGCUGCAUGAGGUCCAUCG ...((((((((((..(((.......)))....))))).((((((.(.....).)))))).((((....))))(((((((.(((((((.....))))))).)))))))..)))))...... ( -56.40) >consensus UAUGCUUCGGCGGCA_________UGCUACUGCCGCCGCCUAGCUGAAAAUCCGGCUGGAGCAUUACAAUCUGCAGCCGCUGGCCAUCGGCCACGGUGACGGGCUGCAUCAGAUCAAGCG ...((...(((((((...............)))))))((((.((((......(((((...((((......))))))))).((((.....))))))))...)))).))............. (-23.12 = -24.88 + 1.75)


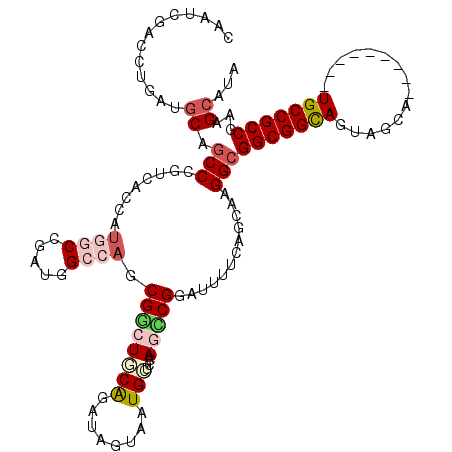
| Location | 18,447,019 – 18,447,130 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.26 |
| Mean single sequence MFE | -45.43 |
| Consensus MFE | -24.38 |
| Energy contribution | -25.58 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.807547 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18447019 111 - 27905053 CAAUCGACCUCAAUCAGCCGGUUACCAUGGCCGAUGGCCAGCGACUGCGCAUCGUGAUGGUGAAUUCGGAUUUUCAGGUAGGCGGCGGCAGUAGCA---------UGCCGCCGAAGCAAA ...((.((((.((....(((((((((((((((...)))).(((....)))......)))))))..))))...)).)))).))((((((((......---------))))))))....... ( -43.00) >DroVir_CAF1 8995 111 - 1 CGCUUGAUUUGAUGCAACCUGUCACUGUGGCAGAUGGCCAACGGCUGCAGAUAGCACUGAUUCAGCCGGAUUUUCAGCAAGGCGGCGGCAGUAGCA---------UGCCGCCGAAGCAUA .((((.......(((.....(((....((((.....)))).(((((((((......)))...))))))))).....)))...((((((((......---------))))))))))))... ( -43.10) >DroGri_CAF1 8803 111 - 1 CAAUGGAUCUAAUGCAGCCCGUCACCGUUGCGGAUGGACUGCGCCUGCAGAUUGCCUUGCUCAAUCCGGAUUUUCAGUUAGGCGGCGGCAGUAGCA---------UGCCGCCGAAGCAUA .............((((.(((((.........))))).))))...(((.....((((.(((.((........)).))).))))(((((((......---------)))))))...))).. ( -39.10) >DroWil_CAF1 3747 111 - 1 CACUCGACCUGAUGCAGCCCGUUACCAUAGCUGAUGGCCAUCGGCUGCAGAUUGUAAUGCGUCAGUCGGAUUUUCAGCCAGGCGGCGGCAAUAUUG---------UGCCGCCGAAGCUCA ...(((((.(((((((...((......((((((((....)))))))).....))...))))))))))))......(((....((((((((......---------))))))))..))).. ( -43.80) >DroMoj_CAF1 7040 111 - 1 CGCUUGAUCUGAUGCAGCCCGUCACAGUGGCCGAUGGGCAGCGGCUUCAAAUAGUACUGAUCCAGCCGGAUUUUCAGCAAGGCGGCGGCAGUAAUA---------UGCCGCCGAAGCAUA .((((.......(((.(((((((.........)))))))..((((((((........)))...)))))........)))...((((((((......---------))))))))))))... ( -45.20) >DroAna_CAF1 3241 120 - 1 CGAUGGACCUCAUGCAGCCGGUGACCAUGGCCGAUGGCCAGCGGCUGCGCAUCGUGAUGCUCCAGCCGGAUUUUCAGGCCGGCGGCGGUAGUGGCAGUAGUGGCCUGCCGCCGAAGCAAA ((((((....)..(((((((.((.((((.....)))).)).))))))).)))))...((((.(.(((((.........)))))((((((((.(.((....)).)))))))))).)))).. ( -58.40) >consensus CAAUCGACCUGAUGCAGCCCGUCACCAUGGCCGAUGGCCAGCGGCUGCAGAUAGUAAUGCUCCAGCCGGAUUUUCAGCAAGGCGGCGGCAGUAGCA_________UGCCGCCGAAGCAUA .............((.(((........((((.....)))).((((((((........)))...)))))............)))(((((((...............)))))))...))... (-24.38 = -25.58 + 1.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:35:04 2006