
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,446,429 – 18,446,549 |
| Length | 120 |
| Max. P | 0.767756 |

| Location | 18,446,429 – 18,446,549 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.83 |
| Mean single sequence MFE | -42.13 |
| Consensus MFE | -27.13 |
| Energy contribution | -26.69 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.40 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.767756 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
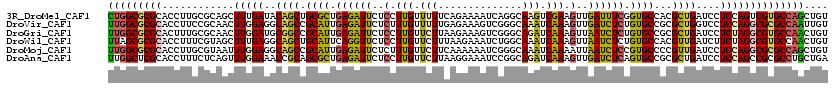
>3R_DroMel_CAF1 18446429 120 + 27905053 CUGGCGCGCACCUUGCGCAGCUUUGAUACAGCUGCGCUGAGAUUCUCCUUGUUUUUCAGAAAAUCAGGCAAGUCGAAGUUGAUUUCGGUGCCACGCUGAUCCUCCAGUCGUGCCAGCUGU ((((((((((((..((((((((.......))))))))...((((..(((.(((((....))))).)))..))))((((....)))))))))...((((......)))).))))))).... ( -46.30) >DroVir_CAF1 8373 120 + 1 UUGGCGCGCACCUUCCGCAACUUGGAGGCAGCCGCAUUGAGAUUCUCUUUGUUUUUGAGAAAGUCGGGCAAAUCAAAGUUGAUCUCUGUGCCGCGCUGGUCCUCCAGGCGCGCCAAUUGU (((((((((.......))..(((((((((((((((((.((((((..(((((..((((.(.....)...)))).)))))..)))))).))).)).))))..)))))))).))))))).... ( -51.90) >DroGri_CAF1 8174 120 + 1 UUGGCGCGCACUUUGCGCAACUUGGAUGCGGCCGCAUUGAGAUUCUCCUUGUUCUUAAGAAAGUCGGGCAGAUCAAAGUUAAUCUCUGUGCCGCGCUGAUCCUCUAGGCGUGCCAACUGU (((((((((.....))((.....((((.(((((((((.((((((..(.(((.(((...(.....)....))).))).)..)))))).))).)).)))))))).....))))))))).... ( -41.20) >DroWil_CAF1 3106 120 + 1 UUAGCGCGCACCUUUCGUAGCUUUGAGGCAGCUGCAUUCAGGUUCUCCUUGUUCUUUAAGAAAUCUGGCAAAUCAAAGUUAAUCUCUGUGCCACGUUGAUCUUCUAGGCGUGCCAACUGU ...((((((((((...((((((.......))))))....))))..........((..(((((((.(((((...((.((.....)).))))))).)))..))))..))))))))....... ( -30.30) >DroMoj_CAF1 6405 120 + 1 UUGGCGCGCACCUUGCGUAAUUUGGAGGCAGCCGCAUUGAGAUUCUCUUUGUUCUUCAAAAAAUCGGGCAAAUCAAAAUUAAUCUCCGUGCCCCGUUGAUCCUCCAGGCGCGCCAGCUGU (((((((((((.....))...(((((((((((.((((.((((((...(((((((...........)))))))........)))))).))))...))))..)))))))))))))))).... ( -41.50) >DroAna_CAF1 2644 120 + 1 UUGGCUCGCACCUUUCUCAGUUUGGAAACCGCAGCGCUGAGAUUCUCCUUGUUCUUAAGGAAAUCCGGCAGAUCAAAGUUGAUCUCAGUGCCGCGCUGAUCCUCCAGCCGCGCCUGCUGA (..((.(((...(((((......)))))..((.(((((((((((..(.(((.(((...((....))...))).))).)..))))))))))).))((((......)))).)))...))..) ( -41.60) >consensus UUGGCGCGCACCUUGCGCAACUUGGAGGCAGCCGCAUUGAGAUUCUCCUUGUUCUUAAGAAAAUCGGGCAAAUCAAAGUUAAUCUCUGUGCCGCGCUGAUCCUCCAGGCGCGCCAACUGU (((((((((............(((((..((((.((((.((((((..(.(((.(((..............))).))).)..)))))).))))...))))....)))))))))))))).... (-27.13 = -26.69 + -0.44)

| Location | 18,446,429 – 18,446,549 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.83 |
| Mean single sequence MFE | -42.67 |
| Consensus MFE | -22.28 |
| Energy contribution | -23.28 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.42 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.616986 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18446429 120 - 27905053 ACAGCUGGCACGACUGGAGGAUCAGCGUGGCACCGAAAUCAACUUCGACUUGCCUGAUUUUCUGAAAAACAAGGAGAAUCUCAGCGCAGCUGUAUCAAAGCUGCGCAAGGUGCGCGCCAG ....((((((((.(((......)))))).((((((((......))).........((((((((.........))))))))...((((((((.......))))))))..)))))..))))) ( -45.90) >DroVir_CAF1 8373 120 - 1 ACAAUUGGCGCGCCUGGAGGACCAGCGCGGCACAGAGAUCAACUUUGAUUUGCCCGACUUUCUCAAAAACAAAGAGAAUCUCAAUGCGGCUGCCUCCAAGUUGCGGAAGGUGCGCGCCAA ....(((((((((.((((((..(((((((((((((((.....)))))...)))).((..(((((.........)))))..))...)).)))))))))).....(....)..))))))))) ( -49.60) >DroGri_CAF1 8174 120 - 1 ACAGUUGGCACGCCUAGAGGAUCAGCGCGGCACAGAGAUUAACUUUGAUCUGCCCGACUUUCUUAAGAACAAGGAGAAUCUCAAUGCGGCCGCAUCCAAGUUGCGCAAAGUGCGCGCCAA ....(((((.........((((..((((.(((..((((((..(((((.(((..............))).)))))..))))))..))).).))))))).....((((.....))))))))) ( -41.24) >DroWil_CAF1 3106 120 - 1 ACAGUUGGCACGCCUAGAAGAUCAACGUGGCACAGAGAUUAACUUUGAUUUGCCAGAUUUCUUAAAGAACAAGGAGAACCUGAAUGCAGCUGCCUCAAAGCUACGAAAGGUGCGCGCUAA ....(((((.(((((..((((......((((((((((.....)))))...)))))....))))..))..........((((...((.((((.......)))).))..))))))).))))) ( -30.20) >DroMoj_CAF1 6405 120 - 1 ACAGCUGGCGCGCCUGGAGGAUCAACGGGGCACGGAGAUUAAUUUUGAUUUGCCCGAUUUUUUGAAGAACAAAGAGAAUCUCAAUGCGGCUGCCUCCAAAUUACGCAAGGUGCGCGCCAA .....((((((((.((((((.....(((((((.(..((......))..).)))))((((((((.........))))))))......))....)))))).....(....)..)))))))). ( -43.90) >DroAna_CAF1 2644 120 - 1 UCAGCAGGCGCGGCUGGAGGAUCAGCGCGGCACUGAGAUCAACUUUGAUCUGCCGGAUUUCCUUAAGAACAAGGAGAAUCUCAGCGCUGCGGUUUCCAAACUGAGAAAGGUGCGAGCCAA ...((....))((((.(.(((..(.((((((.(((((((...(((((.(((...((....))...))).)))))...))))))).)))))).).)))..(((......))).).)))).. ( -45.20) >consensus ACAGCUGGCACGCCUGGAGGAUCAGCGCGGCACAGAGAUCAACUUUGAUUUGCCCGAUUUUCUGAAGAACAAGGAGAAUCUCAAUGCGGCUGCCUCCAAGCUGCGCAAGGUGCGCGCCAA ....(((((.(((((.((((.((....(((((....((((......)))))))))....((((........)))))).))))..(((((((.......)))))))...)).))).))))) (-22.28 = -23.28 + 1.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:35:01 2006