
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,430,514 – 18,430,605 |
| Length | 91 |
| Max. P | 0.964134 |

| Location | 18,430,514 – 18,430,605 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 79.30 |
| Mean single sequence MFE | -17.98 |
| Consensus MFE | -15.54 |
| Energy contribution | -15.27 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.964134 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
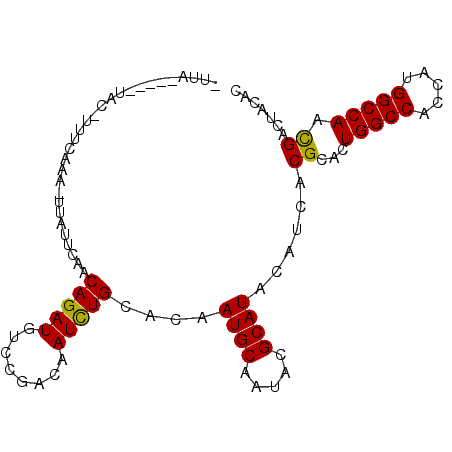
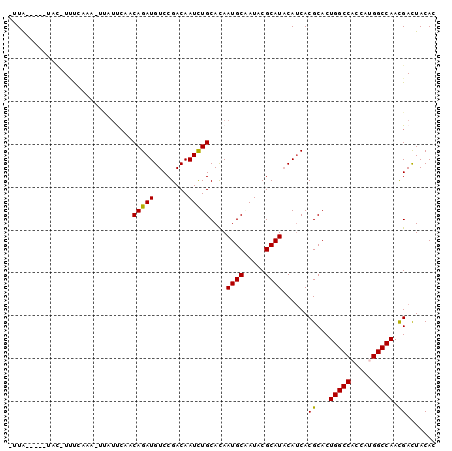
>3R_DroMel_CAF1 18430514 91 + 27905053 -AUA------AC-UUUCAAA-UUAUUCAACAGAUGUCCGACAAUCUGCACAAUGCAAUACGCAUACAUCACGCACUGGCCACCAAGGCCAACGACUACAC -...------..-.......-........(((((........)))))....((((.....))))......((...(((((.....))))).))....... ( -16.00) >DroVir_CAF1 14402 97 + 1 -UUAACCCCUUCGUCUGUUUUCCUUUC--CAGAUGUCCGACAAUUUGCAUAAUGCAAUACGCAUACAUCACGCACUGGCCACCAUGGCCAACGACUACAC -...........(((............--..(((((..(.....((((.....))))....)..)))))......(((((.....)))))..)))..... ( -20.10) >DroGri_CAF1 15294 95 + 1 -UUAA----UUUGUAUAUUCUUUCUCACACAGAUGUCCGACAAUCUGCACAAUGCGAUACGCAUACAUCACGCACUGGCCACUAUGGCCAAUGACUACAC -....----..((((..............(((((........)))))....(((((...)))))........((.(((((.....))))).))..)))). ( -19.20) >DroEre_CAF1 16463 95 + 1 CUUA-C--GUAC-UUUCAAA-UUGUUCAACAGAUGUCCGACAAUCUGCACAAUGCAAUACGCAUACAUCACGCACUGGCCACCAAGGCCAACGACUACAC ....-(--((..-.......-.(((....(((((........)))))....((((.....)))).......))).(((((.....))))))))....... ( -18.00) >DroMoj_CAF1 14004 98 + 1 -UUGACCAACAC-UUUGCUAUAAUUCCAACAGAUGUCCGACAAUCUGCAUAAUGCUAUACGCAUACAUCACGCACUGGCCACUAUGGCCAACGAUUACAC -...........-..(((...........(((((........)))))....((((.....)))).......))).(((((.....))))).......... ( -18.60) >DroAna_CAF1 15923 90 + 1 -CUA------A--UCCCAAU-UCAAUCCACAGAUGUCCGACAAUCUGCACAAUGCCAUUCGCAUUCACCACGCACUGGCCACCAUGGCCAACGACUACAC -...------.--.......-........(((((........)))))...(((((.....))))).....((...(((((.....))))).))....... ( -16.00) >consensus _UUA_____UAC_UUUCAAA_UUAUUCAACAGAUGUCCGACAAUCUGCACAAUGCAAUACGCAUACAUCACGCACUGGCCACCAUGGCCAACGACUACAC .............................(((((........)))))....((((.....))))......((...(((((.....))))).))....... (-15.54 = -15.27 + -0.28)

| Location | 18,430,514 – 18,430,605 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 79.30 |
| Mean single sequence MFE | -27.87 |
| Consensus MFE | -19.86 |
| Energy contribution | -19.70 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.889050 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18430514 91 - 27905053 GUGUAGUCGUUGGCCUUGGUGGCCAGUGCGUGAUGUAUGCGUAUUGCAUUGUGCAGAUUGUCGGACAUCUGUUGAAUAA-UUUGAAA-GU------UAU- .........((((((.....)))))).(((..(((((.......)))))..)))(((((((..(((....)))..))))-)))....-..------...- ( -25.10) >DroVir_CAF1 14402 97 - 1 GUGUAGUCGUUGGCCAUGGUGGCCAGUGCGUGAUGUAUGCGUAUUGCAUUAUGCAAAUUGUCGGACAUCUG--GAAAGGAAAACAGACGAAGGGGUUAA- .....(((.((((((.....))))))(((((((((((.......)))))))))))........))).((((--..........))))(....)......- ( -28.90) >DroGri_CAF1 15294 95 - 1 GUGUAGUCAUUGGCCAUAGUGGCCAGUGCGUGAUGUAUGCGUAUCGCAUUGUGCAGAUUGUCGGACAUCUGUGUGAGAAAGAAUAUACAAA----UUAA- ((((((.((((((((.....)))))))))((((((......)))))).(..(((((((.(.....))))))))..).......)))))...----....- ( -30.10) >DroEre_CAF1 16463 95 - 1 GUGUAGUCGUUGGCCUUGGUGGCCAGUGCGUGAUGUAUGCGUAUUGCAUUGUGCAGAUUGUCGGACAUCUGUUGAACAA-UUUGAAA-GUAC--G-UAAG (((...((.((((((.....)))))).(((..(((((.......)))))..)))(((((((..(((....)))..))))-)))))..-.)))--.-.... ( -28.70) >DroMoj_CAF1 14004 98 - 1 GUGUAAUCGUUGGCCAUAGUGGCCAGUGCGUGAUGUAUGCGUAUAGCAUUAUGCAGAUUGUCGGACAUCUGUUGGAAUUAUAGCAAA-GUGUUGGUCAA- ....((((.((((((.....))))))((((((((((.........))))))))))))))(.(.(((((.(((((......)))))..-))))).).)..- ( -30.50) >DroAna_CAF1 15923 90 - 1 GUGUAGUCGUUGGCCAUGGUGGCCAGUGCGUGGUGAAUGCGAAUGGCAUUGUGCAGAUUGUCGGACAUCUGUGGAUUGA-AUUGGGA--U------UAG- ...(((((.((((((.....))))))((((..(((.((....))..)))..))))((((..(((....)))..))))..-.....))--)------)).- ( -23.90) >consensus GUGUAGUCGUUGGCCAUGGUGGCCAGUGCGUGAUGUAUGCGUAUUGCAUUGUGCAGAUUGUCGGACAUCUGUUGAAAAA_UAUGAAA_GUA_____UAA_ ...(((((.((((((.....))))))((((((((((.........))))))))))))))).(((....)))............................. (-19.86 = -19.70 + -0.16)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:34:55 2006