
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,420,777 – 18,420,870 |
| Length | 93 |
| Max. P | 0.999165 |

| Location | 18,420,777 – 18,420,870 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 70.31 |
| Mean single sequence MFE | -18.72 |
| Consensus MFE | -12.45 |
| Energy contribution | -13.77 |
| Covariance contribution | 1.32 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.66 |
| SVM decision value | 3.41 |
| SVM RNA-class probability | 0.999165 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18420777 93 + 27905053 AUAGCUGGCAAACUUUAUCCCCAAGUCGAGUUACUAAAUUUGCUUGCGAUAUAAUUACCAUACAUUGCCCCACAAAAUAUGCGGCAAAAAUGU .....(((.......((((..((((.((((((....)))))))))).))))......)))....(((((.((.......)).)))))...... ( -15.92) >DroSec_CAF1 6492 80 + 1 -UAGCUGGC------------CAAAUUAAAUUACUAAAUUUGCUUGCGAUAUAAUUACCAUACUUUGCCCCACGAAAUAUGGGGCAACAAUUU -..((.(((------------.(((((.........)))))))).)).................((((((((.......))))))))...... ( -18.70) >DroSim_CAF1 6911 80 + 1 -UAGCUGGC------------CAAAUUAAAUUACUAAAUUUGCUUGCAAUAUAAUUAACAUACUUUGCCCCACGAAAUAUGGGGCAAAAAUUU -..((.(((------------.(((((.........)))))))).))................(((((((((.......)))))))))..... ( -19.80) >DroEre_CAF1 6852 78 + 1 -UAGCUAGCGAAUUU--ACCCCAAGCCAAAUUACUAAAUUUGCUUGCGAUAU------------UUGCUCCACAAAAAGUGGGGCAAAAAUGU -..((.(((((((((--(................)))))))))).))....(------------(((((((((.....))))))))))..... ( -25.69) >DroYak_CAF1 6902 77 + 1 -UAGCUGGCAAACUUUAACCCAAAGUCUAAUUAC---AUUUGCUUGCGAUAU------------AUGGACCACGUAAAAGGGGCCAAAAAUGU -....((((...((((.((.....(((((((..(---(......))..))..------------.)))))...)).))))..))))....... ( -13.50) >consensus _UAGCUGGC_AA_UU___CCCCAAGUCAAAUUACUAAAUUUGCUUGCGAUAUAAUUA_CAUAC_UUGCCCCACGAAAUAUGGGGCAAAAAUGU ...((.((((((..........................)))))).)).................((((((((.......))))))))...... (-12.45 = -13.77 + 1.32)



| Location | 18,420,777 – 18,420,870 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 70.31 |
| Mean single sequence MFE | -20.78 |
| Consensus MFE | -10.23 |
| Energy contribution | -10.75 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.49 |
| SVM decision value | 2.19 |
| SVM RNA-class probability | 0.989886 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18420777 93 - 27905053 ACAUUUUUGCCGCAUAUUUUGUGGGGCAAUGUAUGGUAAUUAUAUCGCAAGCAAAUUUAGUAACUCGACUUGGGGAUAAAGUUUGCCAGCUAU .(((..(((((.(((.....))).)))))...)))...........((..((((((((.((..((......))..)).))))))))..))... ( -23.70) >DroSec_CAF1 6492 80 - 1 AAAUUGUUGCCCCAUAUUUCGUGGGGCAAAGUAUGGUAAUUAUAUCGCAAGCAAAUUUAGUAAUUUAAUUUG------------GCCAGCUA- ......(((((((((.....)))))))))(((.((((........(....)((((((.........))))))------------))))))).- ( -22.70) >DroSim_CAF1 6911 80 - 1 AAAUUUUUGCCCCAUAUUUCGUGGGGCAAAGUAUGUUAAUUAUAUUGCAAGCAAAUUUAGUAAUUUAAUUUG------------GCCAGCUA- ......(((((((((.....)))))))))(((..((((((((.(((((...........))))).))).)))------------))..))).- ( -20.70) >DroEre_CAF1 6852 78 - 1 ACAUUUUUGCCCCACUUUUUGUGGAGCAA------------AUAUCGCAAGCAAAUUUAGUAAUUUGGCUUGGGGU--AAAUUCGCUAGCUA- .....(((((.((((.....)))).))))------------)((((.((((((((((....))))).))))).)))--).............- ( -23.60) >DroYak_CAF1 6902 77 - 1 ACAUUUUUGGCCCCUUUUACGUGGUCCAU------------AUAUCGCAAGCAAAU---GUAAUUAGACUUUGGGUUAAAGUUUGCCAGCUA- ......(((((..((((.(((((((....------------..)))))...((((.---((......)))))).)).))))...)))))...- ( -13.20) >consensus ACAUUUUUGCCCCAUAUUUCGUGGGGCAA_GUAUG_UAAUUAUAUCGCAAGCAAAUUUAGUAAUUUGACUUGGGG___AA_UU_GCCAGCUA_ ......(((((((((.....))))))))).................((..((................................))..))... (-10.23 = -10.75 + 0.52)


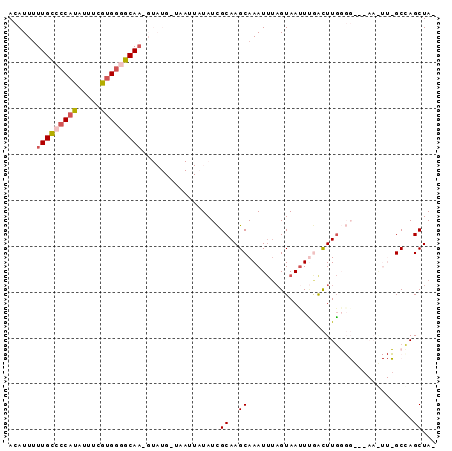
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:34:51 2006