
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,419,362 – 18,419,500 |
| Length | 138 |
| Max. P | 0.869084 |

| Location | 18,419,362 – 18,419,472 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.33 |
| Mean single sequence MFE | -29.66 |
| Consensus MFE | -16.18 |
| Energy contribution | -16.38 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.534922 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18419362 110 + 27905053 GACUACUACUACUAA--AAAAUUUGCUGUAGGUCAGUGCACA--------AGAGAAUCUCUUGAAAGUAGCCAUAGCUAUAAGCUAGCCAACUGGUCAUCACCGCCUCGGCGGCUGACUC (((((...(((((..--.......((((.....))))...((--------((((...))))))..)))))...((((.....))))......))))).(((((((....)))).)))... ( -31.00) >DroSec_CAF1 5114 93 + 1 GACUACUACUACUACUUGAAAUCUGCUGUAGGCCAGU---------------------------AAGUAGCCAUAGCUACAAGCUAGCCAACUGGCCAUCACCGCCUCGGCGGCUGACUC ..............................(((((((---------------------------..(((((....)))))..(....)..))))))).(((((((....)))).)))... ( -27.90) >DroSim_CAF1 5536 93 + 1 AACUACUACUAUUACUUAAAAUCUGCUGUAGUCCAGU---------------------------UAGUAGCCAUAGCUAUAAGCUAGCCAACUGGCCAUCACCGCCUCGGCGGCUGACUC ..(((((((((((((............)))))..)).---------------------------))))))...((((.....))))(((....)))..(((((((....)))).)))... ( -24.40) >DroEre_CAF1 5436 116 + 1 GACUACGACUG---CUGAAAAUCUGCUGCG-GUCAGUGGAAAUGAUGUGCAGAGCACCCCUUGAAAGUAGCCAUAGCUAGGAGCUAGCCAACUGGCCAUCACCUCCUCGGUGGCUGACUC ..(((((((((---(............)))-))).)))).......((((...))))........(((((((((....(((((...(((....)))......)))))..)))))).))). ( -34.70) >DroYak_CAF1 5472 116 + 1 GACUACGACUG---CUGAAAAUCUGAUAUG-GUUAGUGCAAGUUAUGAACAGAGAAUCUUCUGAAAGUAAUCAUAGCUACAAGCUAGCCAACUGGCCAUCACCUCCUCGGUGGCUGACUC ...........---..............((-((((((...((((((((.(((((....))))).......))))))))....))))))))...(((((((........)))))))..... ( -30.30) >consensus GACUACUACUA_UACUGAAAAUCUGCUGUAGGUCAGUG_A__________AGAG_A_C___UGAAAGUAGCCAUAGCUACAAGCUAGCCAACUGGCCAUCACCGCCUCGGCGGCUGACUC ........................((((.....)))).............................(((((....)))))..(((((....)))))..(((((((....)))).)))... (-16.18 = -16.38 + 0.20)


| Location | 18,419,400 – 18,419,500 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.11 |
| Mean single sequence MFE | -29.02 |
| Consensus MFE | -20.85 |
| Energy contribution | -21.13 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.869084 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18419400 100 + 27905053 CA--------AGAGAAUCUCUUGAAAGUAGCCAUAGCUAUAAGCUAGCCAACUGGUCAUCACCGCCUCGGCGGCUGACUCAGAU------------UCAGAUUGAGACUCAGGCAUGCAA ((--------((((...))))))...(((((....)))))..((..(((..(((((((...((((....)))).)))).))).(------------((.....))).....)))..)).. ( -31.80) >DroSec_CAF1 5151 84 + 1 ------------------------AAGUAGCCAUAGCUACAAGCUAGCCAACUGGCCAUCACCGCCUCGGCGGCUGACUCAGAU------------UCAGAUUGAGACUCAGGCAUGCAA ------------------------..((((((.((((.....))))(((....)))..(((((((....)))).)))(((((..------------.....))))).....))).))).. ( -26.60) >DroSim_CAF1 5573 84 + 1 ------------------------UAGUAGCCAUAGCUAUAAGCUAGCCAACUGGCCAUCACCGCCUCGGCGGCUGACUCAGAU------------UCAGAUUGAGACUCAGGCAUGCAA ------------------------..((((((.((((.....))))(((....)))..(((((((....)))).)))(((((..------------.....))))).....))).))).. ( -26.60) >DroEre_CAF1 5472 108 + 1 AAUGAUGUGCAGAGCACCCCUUGAAAGUAGCCAUAGCUAGGAGCUAGCCAACUGGCCAUCACCUCCUCGGUGGCUGACUCAGAU------------UCAGAUUGAGACUCAGGCAUGCAA .............(((..(((.((...(((((((....(((((...(((....)))......)))))..))))))).(((((..------------.....)))))..)))))..))).. ( -29.80) >DroYak_CAF1 5508 120 + 1 AGUUAUGAACAGAGAAUCUUCUGAAAGUAAUCAUAGCUACAAGCUAGCCAACUGGCCAUCACCUCCUCGGUGGCUGACUCAGAUUCAGAUUCAGAUUCAGAUUGAGACUCAGGCAUGCAA ((((((((.(((((....))))).......))))))))....((..(((....(((((((........)))))))..(((((..((.((.......)).))))))).....)))..)).. ( -30.30) >consensus __________AGAG_A_C___UGAAAGUAGCCAUAGCUACAAGCUAGCCAACUGGCCAUCACCGCCUCGGCGGCUGACUCAGAU____________UCAGAUUGAGACUCAGGCAUGCAA ..........................((((((.((((.....))))(((....)))..(((((((....)))).)))(((((...................))))).....))).))).. (-20.85 = -21.13 + 0.28)

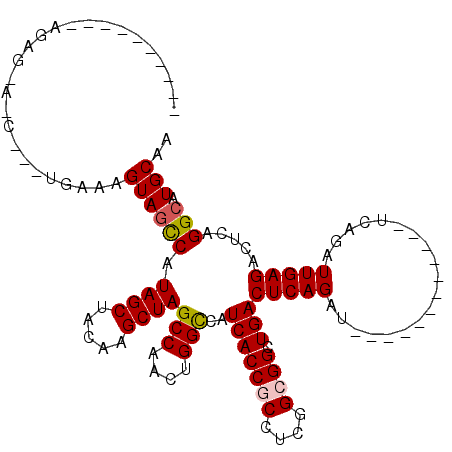
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:34:49 2006