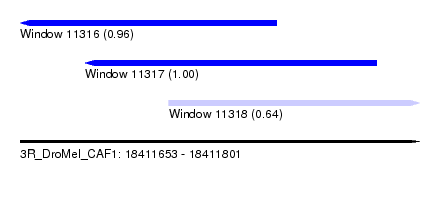
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,411,653 – 18,411,801 |
| Length | 148 |
| Max. P | 0.998644 |

| Location | 18,411,653 – 18,411,748 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 78.34 |
| Mean single sequence MFE | -36.88 |
| Consensus MFE | -19.64 |
| Energy contribution | -20.92 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.53 |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.958638 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18411653 95 - 27905053 UUGGCCAGCGACUUGGCUGCGGCCAGCUGCAACUUCUGGGUGGCGGCAUUCUU-GUUGCUCUUAGCCGGCGGAGCUAUUUUCU---UCUUCUGCACACC .((((.((((((..(..(((.((((.((.........)).)))).)))..)..-))))))....))))((((((.........---..))))))..... ( -33.60) >DroVir_CAF1 4773 95 - 1 UUGGCCAACUGCUUGGCCGCGGCCAGCUGCAGCGUUUUCGUGGCCGCAUUUUU-GGUCACUUUGGACGCUGCAGUGGGCUUCUUUUU---CUGCACACC ..((((((....))))))(.((((.((((((((((((..(((((((......)-))))))...)))))))))))).)))).).....---......... ( -47.00) >DroSim_CAF1 5029 95 - 1 UUGGCCAGCGACUUGGCUGCGGCCAGCUGCAGCUUCUGGGUGGCGGCAUUCUU-GUUGCUCUUGGCCGGCAGAGCUACUUUCU---UCUUCUGCACACC .((((((((.((((((((((((....))))))))...)))).))((((.....-..))))..))))))((((((.........---..))))))..... ( -38.80) >DroEre_CAF1 4993 95 - 1 UUGGCCAGCGAUUUGGCUGCGGCCAGCUGCAGUUUCUGGGUGGCGACAUUCUU-GCUGCUCUUCGCAGCCGGAGCUAUUUUCC---UUUUCUGCACGCC ..(((..(((....((((((((..((..(((((....(((((....)))))..-))))).))))))))))((((.....))))---.....)))..))) ( -35.90) >DroYak_CAF1 4966 95 - 1 UUGGCCACCGACUUGGCUGCGGCCAGCUGCAGCUUCUGGGUGGCGACAUUCUU-GCUGCUUUUGGCCACCGGAGCUAUUUUCU---UUUUCUGCACGCC ..(((((......)))))((((((((..(((((....(((((....)))))..-)))))..))))))..(((((.........---..)))))...)). ( -35.50) >DroAna_CAF1 5164 95 - 1 UUAGCCAGCUUUUUUGCUGCGGCCAGCUGCAGUUUCUGGGUGGCAUCAUUCUUAGGUGCU-UUAACUGCUGGAGCCACCUUUU---UCUUCUGGAUACC ...((((.((.....(((((((....)))))))....)).))))(((......(((((((-(((.....))))).)))))...---.......)))... ( -30.49) >consensus UUGGCCAGCGACUUGGCUGCGGCCAGCUGCAGCUUCUGGGUGGCGGCAUUCUU_GCUGCUCUUGGCCGCCGGAGCUAUUUUCU___UCUUCUGCACACC ...((((.(.(...((((((((....))))))))..).).)))).............(((((........)))))........................ (-19.64 = -20.92 + 1.28)


| Location | 18,411,677 – 18,411,785 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 67.35 |
| Mean single sequence MFE | -39.10 |
| Consensus MFE | -25.76 |
| Energy contribution | -23.33 |
| Covariance contribution | -2.42 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.66 |
| SVM decision value | 3.17 |
| SVM RNA-class probability | 0.998644 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18411677 108 - 27905053 CUACUUGGAUUUCUUGGCCUUUCCCUUGGGCAAGGGUUUGGCCAGCGACUUGGCUGCGGCCAGCUGCAACUUCUGGGUGGCGGCAUUCUUGUUGCUCUUAGCCGGCGG-------- ....((((......(((((...(((((....)))))...)))))(((((..(..(((.((((.((.........)).)))).)))..)..)))))......))))...-------- ( -42.30) >DroWil_CAF1 12479 113 - 1 CUUCUUAAGCUUCUUUGCUGUUACUUUCUUUAGAGGCUGGGCCAACUCCUUCGCAGCAGUCAAUUUUAAUGUCUGGGUGG---CCUUCUUGUUGAUCUUAUUUGACGAGGAUUUCG .......(((((((.................))))))).(((((...(((..(((..((......))..)))..))))))---))(((((((..(......)..)))))))..... ( -28.03) >DroYak_CAF1 4990 108 - 1 CUACUUGGAUUUCUUGGCCUUUCCCUUGGGCAAGGGUUUGGCCACCGACUUGGCUGCGGCCAGCUGCAGCUUCUGGGUGGCGACAUUCUUGCUGCUUUUGGCCACCGG-------- .....(((......(((((.........(((((((((.(.((((((.(...((((((((....))))))))..).)))))).).)))))))))......)))))))).-------- ( -46.96) >consensus CUACUUGGAUUUCUUGGCCUUUCCCUUGGGCAAGGGUUUGGCCAACGACUUGGCUGCGGCCAGCUGCAACUUCUGGGUGGCG_CAUUCUUGUUGCUCUUAGCCGACGG________ .....(((......(((((...(((((....)))))...))))).....(((((....)))))..(((((....(((((....)))))..)))))......)))............ (-25.76 = -23.33 + -2.42)

| Location | 18,411,708 – 18,411,801 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 76.78 |
| Mean single sequence MFE | -26.73 |
| Consensus MFE | -12.07 |
| Energy contribution | -11.85 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.45 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.635737 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18411708 93 + 27905053 CCCAGAAGUUGCAGCUGGCCGCAGCCAAGUCGCUGGCCAAACCCUUGCCCAAGGGAAAGGCCAAGAAAUCCAA---GUAGUAG-CUUAAAAUACAU-U .....(((((((.((((((....)))...((..(((((...(((((....)))))...))))).))......)---)).))))-))).........-. ( -30.20) >DroPse_CAF1 6268 95 + 1 CACAGAAGCUGCAGCUGGCCGCAGCCAAGACGCUGGCCAAGCCGCUGCAGAAGGUCAAGUCCAAGAAGUUGAA---GAAAUAGUCGUAAACUACAUUU ....((..(((((((.(((....((((......))))...))))))))))....))........(.((((...---((.....))...)))).).... ( -27.10) >DroSec_CAF1 5079 93 + 1 CCCAGAAGCUGCAGCUGGCCGCAGCCAAGUCGCUGGCCAAGCCCUUGCCCAAGGGAAAGGCCAAGAAAUCCAA---GUAGUAG-CUUAAAAUACAU-U .....(((((((.((((((....)))...((..(((((...(((((....)))))...))))).))......)---)).))))-))).........-. ( -33.70) >DroMoj_CAF1 4261 95 + 1 CAAAGACACUGCAGCUGGCCGCAGCCAAACAACUGGCCAAGCCAUUGCAGAAGAAGAUUGGCAAAAAGCUAAA---GAAGUAGAAUUGUAGUAUAUUU .....(((((((((.((((....((((......))))...)))))))))).......(((((.....))))).---..........)))......... ( -23.90) >DroAna_CAF1 5219 93 + 1 CCCAGAAACUGCAGCUGGCCGCAGCAAAAAAGCUGGCUAAACCCCUGCCCAAG---AAAGCAAAGAAGUCCAAGAAGUAGUAG-UUUAAAAUGUAU-C .....(((((((.(((((((..(((......))))))).......(((.....---...))).............))).))))-))).........-. ( -18.40) >DroPer_CAF1 6333 95 + 1 CACAGAAGCUGCAGCUGGCCGCAGCCAAGACGCUGGCCAAGCCGCUGCAGAAGGUCAAGUCCAAGAAGUUGAA---GAAAUAGUCGUAAACUACAUUU ....((..(((((((.(((....((((......))))...))))))))))....))........(.((((...---((.....))...)))).).... ( -27.10) >consensus CACAGAAGCUGCAGCUGGCCGCAGCCAAGACGCUGGCCAAGCCCCUGCACAAGGGAAAGGCCAAGAAGUCCAA___GAAGUAG_CUUAAAAUACAU_U ..........((((.(((((..(((......)))))))).....)))).................................................. (-12.07 = -11.85 + -0.22)

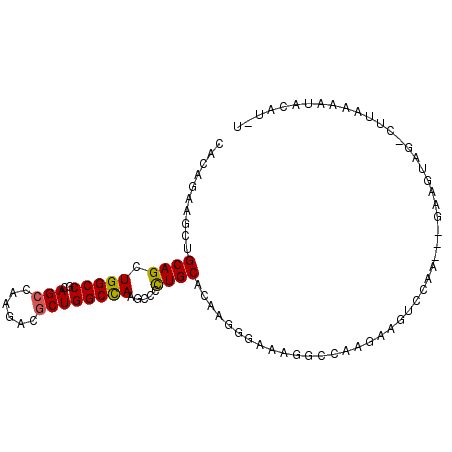
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:34:43 2006