
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,388,974 – 18,389,080 |
| Length | 106 |
| Max. P | 0.919810 |

| Location | 18,388,974 – 18,389,080 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 82.55 |
| Mean single sequence MFE | -13.03 |
| Consensus MFE | -8.85 |
| Energy contribution | -9.48 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.68 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
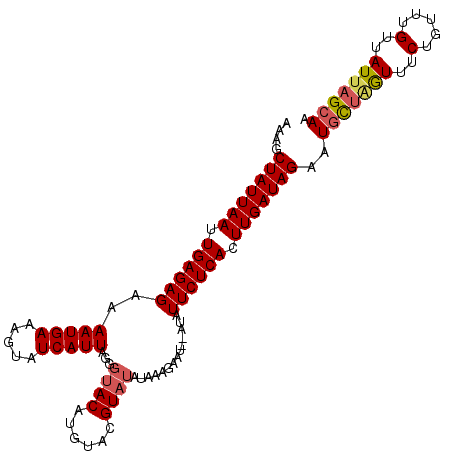
>3R_DroMel_CAF1 18388974 106 + 27905053 UUGCUAAUAACAAACAGAAACUAACAUUCUAUCAAAUGAGAAUAU--AUUCUACAUAUACGUAUGUGUACCCUAAUGAUACUUUCAUUUUCUCUCAAUUAAUAGCUUU ..((((.(((.....((((.......))))......(((((....--....(((((((....)))))))....(((((.....)))))...))))).))).))))... ( -15.80) >DroSec_CAF1 8586 89 + 1 UUGCUAAUAACAAACAGAAACCAACAUUCUAUCAAAUGAGAAUAU--AUUC-----------------UUCCUAAUGAUACUUUCAUUUUCUCUCAAUUAAUAGCUUU ..((((.(((.....((((......(((((........)))))..--....-----------------.....(((((.....))))))))).....))).))))... ( -9.70) >DroEre_CAF1 10594 108 + 1 UUGCAAAUAACAAACAGAAAUCGGCAUUCUAUCAAGUGAGAAUAUAUAUAUUUUAUAUACGUACAUGUACCCUAAUGAUACUUUCAUUUUCUCUCAAUUAAUAGCUUU ..((...........((((.......)))).....(.(((((((((((.....)))))).((((((........))).))).......))))).)........))... ( -12.20) >DroYak_CAF1 9514 106 + 1 UUGCUUAUAGCAAACAGAAACUAGAAUUCUAUCAAGUGAGAAUAU--AUUCUUUAUAUACGUACACGUACCCUAAUGAUACUUUCAUUUUCUCUCAAUUAAUAGCUUC ((((.....))))...............((((.((.(((((....--..........((((....))))....(((((.....)))))...))))).)).)))).... ( -14.40) >consensus UUGCUAAUAACAAACAGAAACCAACAUUCUAUCAAAUGAGAAUAU__AUUCUUUAUAUACGUACAUGUACCCUAAUGAUACUUUCAUUUUCUCUCAAUUAAUAGCUUU ..((((.(((.....((((.......))))......(((((................((((....))))....(((((.....)))))...))))).))).))))... ( -8.85 = -9.48 + 0.62)



| Location | 18,388,974 – 18,389,080 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 82.55 |
| Mean single sequence MFE | -21.45 |
| Consensus MFE | -17.39 |
| Energy contribution | -18.20 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.919810 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18388974 106 - 27905053 AAAGCUAUUAAUUGAGAGAAAAUGAAAGUAUCAUUAGGGUACACAUACGUAUAUGUAGAAU--AUAUUCUCAUUUGAUAGAAUGUUAGUUUCUGUUUGUUAUUAGCAA ....(((((((.((((((.........(((((.....)))))(((((....))))).....--...)))))).)))))))..(((((((..(.....)..))))))). ( -24.40) >DroSec_CAF1 8586 89 - 1 AAAGCUAUUAAUUGAGAGAAAAUGAAAGUAUCAUUAGGAA-----------------GAAU--AUAUUCUCAUUUGAUAGAAUGUUGGUUUCUGUUUGUUAUUAGCAA ...((((.((((.((.(((((..((...(((((...((((-----------------....--...))))....))))).....))..))))).)).)))).)))).. ( -17.40) >DroEre_CAF1 10594 108 - 1 AAAGCUAUUAAUUGAGAGAAAAUGAAAGUAUCAUUAGGGUACAUGUACGUAUAUAAAAUAUAUAUAUUCUCACUUGAUAGAAUGCCGAUUUCUGUUUGUUAUUUGCAA ....(((((((.((((((.........(((((.....)))))......(((((((...))))))).)))))).)))))))..(((.(((..(.....)..))).))). ( -22.30) >DroYak_CAF1 9514 106 - 1 GAAGCUAUUAAUUGAGAGAAAAUGAAAGUAUCAUUAGGGUACGUGUACGUAUAUAAAGAAU--AUAUUCUCACUUGAUAGAAUUCUAGUUUCUGUUUGCUAUAAGCAA (((.(((((((.((((((..(((((.....)))))...(((((....))))).........--...)))))).)))))))..)))..........((((.....)))) ( -21.70) >consensus AAAGCUAUUAAUUGAGAGAAAAUGAAAGUAUCAUUAGGGUACAUGUACGUAUAUAAAGAAU__AUAUUCUCACUUGAUAGAAUGCUAGUUUCUGUUUGUUAUUAGCAA ....(((((((.((((((..(((((.....)))))...((((......))))..............)))))).)))))))..(((((((..(.....)..))))))). (-17.39 = -18.20 + 0.81)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:34:35 2006