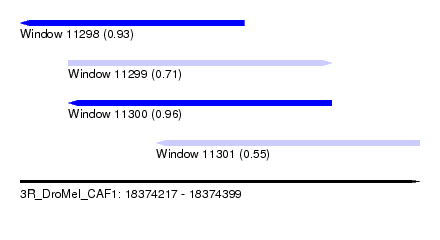
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,374,217 – 18,374,399 |
| Length | 182 |
| Max. P | 0.958151 |

| Location | 18,374,217 – 18,374,319 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 93.92 |
| Mean single sequence MFE | -21.89 |
| Consensus MFE | -16.56 |
| Energy contribution | -17.44 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.930510 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18374217 102 - 27905053 AAGGCUGGUGAUAUUUCCUCACUAGCCAAACACCAUUUUGUCUUCCCAAUUAAGUUUCUUUAUUAUUAGAAUUGUUAUUUGCCUACAUUUGUGGGUUUUUAU ..(((((((((.......)))))))))((((.((((..(((.....(((.(((..((((........))))...))).)))...)))...)))))))).... ( -24.70) >DroSec_CAF1 38691 102 - 1 AUGGCUGGUGAUAUUUCCUCACUAGCCAAACACCAUUUUAUCUUCCCAAUUAAGAUUCUUUAUUAUUAGAAUUGUUAUUUGCCUACAUUUGUGGGUUUUUAU .((((((((((.......)))))))))).........................((((((........)))))).......((((((....))))))...... ( -26.30) >DroSim_CAF1 39981 102 - 1 AUGGCUGGUGAUAUUUCCUCACUAGCCAAACACCAUUUUGUCUUCCCAAUUAAGUUUCUUUAUUAUUAGAAUUGUUAUUUGCCUACAUUUGUGGGUUUUUAU .((((((((((.......))))))))))(((.((((..(((.....(((.(((..((((........))))...))).)))...)))...)))))))..... ( -24.60) >DroEre_CAF1 39706 102 - 1 AUGGCUGGUGAUAUUUCCUCACUAGCAAAAUACCACUUUGUCUGCCCAAUUAAGUUUCUCUAUUAUUAGAAUUGUUAUUUGCCUACAUUUGUGAGUUUUUAU ...((((((((.......)))))))).(((.(((((..(((..((.......(((...((((....))))))).......))..)))...))).)).))).. ( -19.14) >DroYak_CAF1 39705 102 - 1 AUGGCUGGCGAUAUUUCCUCACUAGCAAAAUACCAUUUAGUCUUCCCAAUUAAGUUUCUUUAUUAUUAGAAUUGUUAUUUGUCUACAUUGGUGAAUUUUUAU ...(((((.((.......)).)))))....(((((..(((......(((.(((..((((........))))...))).))).)))...)))))......... ( -14.70) >consensus AUGGCUGGUGAUAUUUCCUCACUAGCCAAACACCAUUUUGUCUUCCCAAUUAAGUUUCUUUAUUAUUAGAAUUGUUAUUUGCCUACAUUUGUGGGUUUUUAU .((((((((((.......))))))))))...........................((((........)))).........((((((....))))))...... (-16.56 = -17.44 + 0.88)

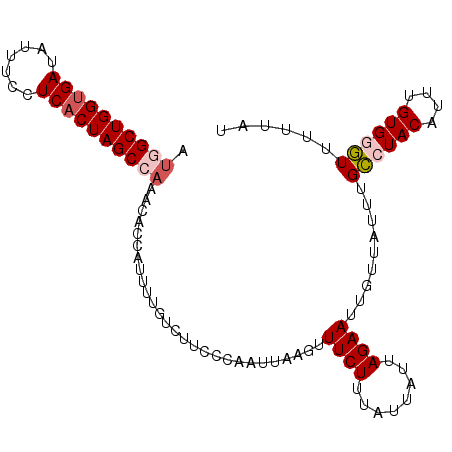
| Location | 18,374,239 – 18,374,359 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.33 |
| Mean single sequence MFE | -26.73 |
| Consensus MFE | -20.54 |
| Energy contribution | -21.18 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.714925 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18374239 120 + 27905053 AAAUAACAAUUCUAAUAAUAAAGAAACUUAAUUGGGAAGACAAAAUGGUGUUUGGCUAGUGAGGAAAUAUCACCAGCCUUGCAGGCCCAAUAAAGAUUCACUUGUUUGGCAAGCUCUUAA ......................(((.(((.((((((....(.....).(((..((((.((((.......)))).))))..)))..)))))).))).))).((((.....))))....... ( -27.80) >DroSec_CAF1 38713 120 + 1 AAAUAACAAUUCUAAUAAUAAAGAAUCUUAAUUGGGAAGAUAAAAUGGUGUUUGGCUAGUGAGGAAAUAUCACCAGCCAUGCAGGCCCAAUAAAGAUUCACUCGUUCGGCAAGCUCCUAA ......................(((((((.((((((............(((.(((((.((((.......)))).))))).)))..)))))).)))))))..................... ( -29.84) >DroSim_CAF1 40003 120 + 1 AAAUAACAAUUCUAAUAAUAAAGAAACUUAAUUGGGAAGACAAAAUGGUGUUUGGCUAGUGAGGAAAUAUCACCAGCCAUGCAGGCCCAAUAAAGAUUCACUUGUUUGGCAAGCUCCUAA ......................(((.(((.((((((....(.....).(((.(((((.((((.......)))).))))).)))..)))))).))).))).((((.....))))....... ( -28.50) >DroEre_CAF1 39728 120 + 1 AAAUAACAAUUCUAAUAAUAGAGAAACUUAAUUGGGCAGACAAAGUGGUAUUUUGCUAGUGAGGAAAUAUCACCAGCCAUGCAGGCCCAAUAAAGAUUCACUUGUUUGGCAGCCUCCUAA ....(((((.((((....))))(((.(((.(((((((.......((((((((((.((....))))))))))))..((...))..))))))).))).)))..))))).............. ( -27.20) >DroYak_CAF1 39727 120 + 1 AAAUAACAAUUCUAAUAAUAAAGAAACUUAAUUGGGAAGACUAAAUGGUAUUUUGCUAGUGAGGAAAUAUCGCCAGCCAUGCAGGCCCAAUAAAGAUUCACUUGUUUGGCAACCUCUUAA ......................(((.(((.((((((....((..(((((....((...((((.......)))))))))))..)).)))))).))).)))........(....)....... ( -20.30) >consensus AAAUAACAAUUCUAAUAAUAAAGAAACUUAAUUGGGAAGACAAAAUGGUGUUUGGCUAGUGAGGAAAUAUCACCAGCCAUGCAGGCCCAAUAAAGAUUCACUUGUUUGGCAAGCUCCUAA ......................(((.(((.((((((...........((((..((((.((((.......)))).))))))))...)))))).))).))).((((.....))))....... (-20.54 = -21.18 + 0.64)


| Location | 18,374,239 – 18,374,359 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.33 |
| Mean single sequence MFE | -29.06 |
| Consensus MFE | -23.00 |
| Energy contribution | -23.88 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.958151 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18374239 120 - 27905053 UUAAGAGCUUGCCAAACAAGUGAAUCUUUAUUGGGCCUGCAAGGCUGGUGAUAUUUCCUCACUAGCCAAACACCAUUUUGUCUUCCCAAUUAAGUUUCUUUAUUAUUAGAAUUGUUAUUU ......(((((.....)))))(((.(((.((((((...(((((((((((((.......)))))))))..........))))...)))))).))).)))...................... ( -29.40) >DroSec_CAF1 38713 120 - 1 UUAGGAGCUUGCCGAACGAGUGAAUCUUUAUUGGGCCUGCAUGGCUGGUGAUAUUUCCUCACUAGCCAAACACCAUUUUAUCUUCCCAAUUAAGAUUCUUUAUUAUUAGAAUUGUUAUUU ......(((((.....)))))(((((((.((((((..((..((((((((((.......))))))))))..))............)))))).)))))))...................... ( -31.94) >DroSim_CAF1 40003 120 - 1 UUAGGAGCUUGCCAAACAAGUGAAUCUUUAUUGGGCCUGCAUGGCUGGUGAUAUUUCCUCACUAGCCAAACACCAUUUUGUCUUCCCAAUUAAGUUUCUUUAUUAUUAGAAUUGUUAUUU ......(((((.....)))))(((.(((.((((((...(((((((((((((.......))))))))))..........)))...)))))).))).)))...................... ( -30.10) >DroEre_CAF1 39728 120 - 1 UUAGGAGGCUGCCAAACAAGUGAAUCUUUAUUGGGCCUGCAUGGCUGGUGAUAUUUCCUCACUAGCAAAAUACCACUUUGUCUGCCCAAUUAAGUUUCUCUAUUAUUAGAAUUGUUAUUU ...((......)).(((((..(((.(((.(((((((..(((..((((((((.......))))))))............)))..))))))).))).)))((((....)))).))))).... ( -31.94) >DroYak_CAF1 39727 120 - 1 UUAAGAGGUUGCCAAACAAGUGAAUCUUUAUUGGGCCUGCAUGGCUGGCGAUAUUUCCUCACUAGCAAAAUACCAUUUAGUCUUCCCAAUUAAGUUUCUUUAUUAUUAGAAUUGUUAUUU ....(((((((((.....(((((....))))).((((.....))))))))).....))))..((((((......(((((((.......)))))))((((........))))))))))... ( -21.90) >consensus UUAGGAGCUUGCCAAACAAGUGAAUCUUUAUUGGGCCUGCAUGGCUGGUGAUAUUUCCUCACUAGCCAAACACCAUUUUGUCUUCCCAAUUAAGUUUCUUUAUUAUUAGAAUUGUUAUUU ..............(((((..(((.(((.((((((...(((((((((((((.......))))))))))..........)))...)))))).))).)))((((....)))).))))).... (-23.00 = -23.88 + 0.88)

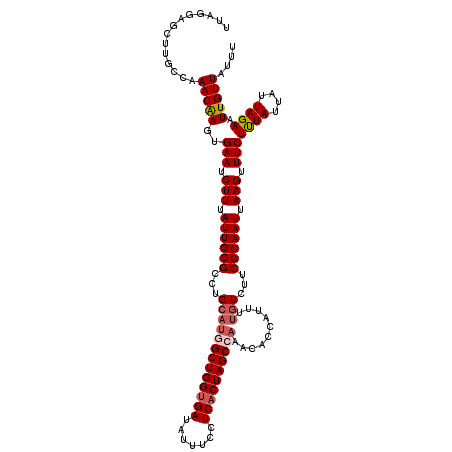

| Location | 18,374,279 – 18,374,399 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.17 |
| Mean single sequence MFE | -31.84 |
| Consensus MFE | -24.38 |
| Energy contribution | -25.38 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.546435 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18374279 120 - 27905053 UAUUCUAAGGGCGCUAUUAAAACGUAGUUAAAUUGCGCUAUUAAGAGCUUGCCAAACAAGUGAAUCUUUAUUGGGCCUGCAAGGCUGGUGAUAUUUCCUCACUAGCCAAACACCAUUUUG ........(((((((.((((..((((((...))))))...)))).)))...((((..(((.....)))..))))))))....(((((((((.......)))))))))............. ( -31.70) >DroSec_CAF1 38753 120 - 1 UAUUCUAAGGGCGCUAUUAAAACGUAGUUAAAUUGCGCUAUUAGGAGCUUGCCGAACGAGUGAAUCUUUAUUGGGCCUGCAUGGCUGGUGAUAUUUCCUCACUAGCCAAACACCAUUUUA ..((((((.(((((..((((.......))))...))))).))))))((..(((.((..((......))..)).)))..)).((((((((((.......))))))))))............ ( -37.40) >DroSim_CAF1 40043 120 - 1 UAUUCUAAGGGCGAUAUUAAAACGUAGUUAAAUUGCGCUAUUAGGAGCUUGCCAAACAAGUGAAUCUUUAUUGGGCCUGCAUGGCUGGUGAUAUUUCCUCACUAGCCAAACACCAUUUUG ..((((((.((((...((((.......))))....)))).))))))((..(((.....(((((....))))).)))..)).((((((((((.......))))))))))............ ( -32.80) >DroEre_CAF1 39768 120 - 1 UAUUCAAAGGGCGCUAUUAAAACGUAGUUAAAUUGCGCUAUUAGGAGGCUGCCAAACAAGUGAAUCUUUAUUGGGCCUGCAUGGCUGGUGAUAUUUCCUCACUAGCAAAAUACCACUUUG ....((((((((((..((((.......))))...)))))....((((((..((((..(((.....)))..)))))))).....((((((((.......))))))))......)).))))) ( -32.40) >DroYak_CAF1 39767 120 - 1 UAUUCAAAGGGCGCCAUUAAAACGUAGUUAAAUUGCGCUAUUAAGAGGUUGCCAAACAAGUGAAUCUUUAUUGGGCCUGCAUGGCUGGCGAUAUUUCCUCACUAGCAAAAUACCAUUUAG .........((((((.((((..((((((...))))))...))))..)).)))).....(((((.....(((((((((.....))))..))))).....)))))................. ( -24.90) >consensus UAUUCUAAGGGCGCUAUUAAAACGUAGUUAAAUUGCGCUAUUAGGAGCUUGCCAAACAAGUGAAUCUUUAUUGGGCCUGCAUGGCUGGUGAUAUUUCCUCACUAGCCAAACACCAUUUUG ........((((..........((((((...))))))(((..(((((((((.....)))))...))))...)))))))...((((((((((.......))))))))))............ (-24.38 = -25.38 + 1.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:34:27 2006