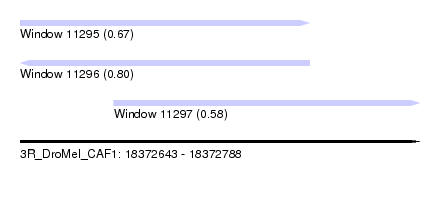
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,372,643 – 18,372,788 |
| Length | 145 |
| Max. P | 0.797019 |

| Location | 18,372,643 – 18,372,748 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 79.73 |
| Mean single sequence MFE | -45.68 |
| Consensus MFE | -26.53 |
| Energy contribution | -27.93 |
| Covariance contribution | 1.41 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.672545 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
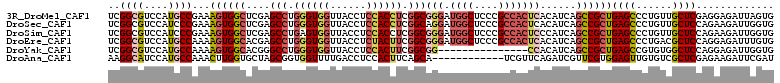
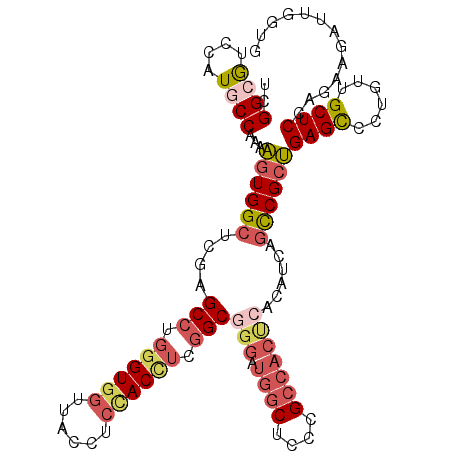
>3R_DroMel_CAF1 18372643 105 + 27905053 GGUGGUGGUCACCGCCUCGCCGGUAGCAUGUCCUUCGGCGUCCAUGCCGAAAGUGGCUCGAGCCUGGGUGGUUACCUCCACCUCGGCGGGAUGGCUCCCGCCACU (((((.(((.(((((((.((((..(((....(.(((((((....))))))).)..))))).))..))))))).))).)))))..(((((((....)))))))... ( -56.50) >DroSec_CAF1 37129 105 + 1 GGUGGUGGUCACAGCCUCGCCGGUAGCGUGUCCUUCGGCGUCCAUCCCGAAAGUGGCUCGAGCCUGGGUGGUUACCUCCACCUCGGCAGGAUGGCUCCCGCCACU ((((((((....((((..(((((.((......)))))))((((..(((....).)).....(((.((((((......)))))).))).)))))))).)))))))) ( -49.70) >DroSim_CAF1 38410 105 + 1 GGUGGUGGUCACAGCCUCGCCGGUAGCGUGUCCUUCGGCGUCCAUCCCGAAAGUGGCUCGAGCCUGAGUGGUUACCUCCACCUCGGCGGGAUGGCUCCCGCCACU (((((.(((...((((.(.(.(((..((.(((((((((.(.....)))).))).))).)).))).).).))))))).)))))..(((((((....)))))))... ( -47.20) >DroEre_CAF1 38152 105 + 1 GGUGGUGGUCACUGCCUCCUCGGUAGCAUGUCCUUCGGCGUCCAUGCCAAAAGUGGCACGAGCCUGGGUGGUUACCUCUACUUCGGCGGGAUGGCUCCCGCCACU (((((.(((.((..(((.((((...(.(((((....))))).).(((((....)))))))))...)))..)).))).)))))..(((((((....)))))))... ( -47.40) >DroYak_CAF1 38100 90 + 1 AGUGGUGGUCACUGCCUCCCCGGUAGCAUGUCCUUCGGCGUCCAUGCCAAAAGUGGCACGGGCCUGGGUGGUUACCUCCACUUCGGCGG---------------C (((((.(((.((..(((.((((...(.(((((....))))).).(((((....)))))))))...)))..)).))).))))).......---------------. ( -37.00) >DroAna_CAF1 48837 93 + 1 GGUGGUGGUCACCGCGUCGGCGGUAGCAUGACCCAAGGCAUCCAUGCCAAACUUGGUGCUAGCGGUGGUUUUGACCUCCACUUCAGCA------------UCGUU (((.(((.(.(((((....)))))).))).)))...((((....))))......((((((((.((.(((....))).)).))..))))------------))... ( -36.30) >consensus GGUGGUGGUCACAGCCUCGCCGGUAGCAUGUCCUUCGGCGUCCAUGCCAAAAGUGGCUCGAGCCUGGGUGGUUACCUCCACCUCGGCGGGAUGGCUCCCGCCACU (((((.(((.((.((((....((((..(((((....)))))...))))......(((....))).)))).)).))).)))))..(((.(((....))).)))... (-26.53 = -27.93 + 1.41)

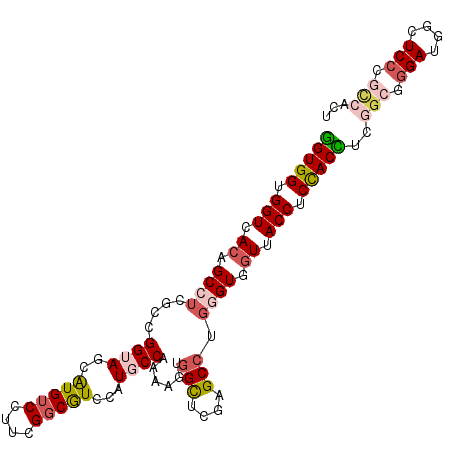
| Location | 18,372,643 – 18,372,748 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 79.73 |
| Mean single sequence MFE | -42.85 |
| Consensus MFE | -29.82 |
| Energy contribution | -32.53 |
| Covariance contribution | 2.71 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.797019 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18372643 105 - 27905053 AGUGGCGGGAGCCAUCCCGCCGAGGUGGAGGUAACCACCCAGGCUCGAGCCACUUUCGGCAUGGACGCCGAAGGACAUGCUACCGGCGAGGCGGUGACCACCACC ..((((((((....)))))))).(((((.(((.(((.(((.(((....))).((((((((......))))))))....((.....))).)).))).))).))))) ( -54.50) >DroSec_CAF1 37129 105 - 1 AGUGGCGGGAGCCAUCCUGCCGAGGUGGAGGUAACCACCCAGGCUCGAGCCACUUUCGGGAUGGACGCCGAAGGACACGCUACCGGCGAGGCUGUGACCACCACC ..((((((((....)))))))).(((((.(((....)))..((.(((((((.(((((((........)))))))...(((.....))).)))).))))).))))) ( -46.30) >DroSim_CAF1 38410 105 - 1 AGUGGCGGGAGCCAUCCCGCCGAGGUGGAGGUAACCACUCAGGCUCGAGCCACUUUCGGGAUGGACGCCGAAGGACACGCUACCGGCGAGGCUGUGACCACCACC ..((((((((....)))))))).(((((.(((.((..(((.(((....))).(((((((........)))))))....((.....)))))...)).))).))))) ( -47.90) >DroEre_CAF1 38152 105 - 1 AGUGGCGGGAGCCAUCCCGCCGAAGUAGAGGUAACCACCCAGGCUCGUGCCACUUUUGGCAUGGACGCCGAAGGACAUGCUACCGAGGAGGCAGUGACCACCACC ..((((((((....)))))))).....(.(((....)))).((.(((((((.(((((((((((..(......)..)))))))..)))).)))).)))))...... ( -41.90) >DroYak_CAF1 38100 90 - 1 G---------------CCGCCGAAGUGGAGGUAACCACCCAGGCCCGUGCCACUUUUGGCAUGGACGCCGAAGGACAUGCUACCGGGGAGGCAGUGACCACCACU .---------------.......(((((.(((..((.(((.(((((((((((....))))))))..)))...((........)).))).)).....))).))))) ( -36.00) >DroAna_CAF1 48837 93 - 1 AACGA------------UGCUGAAGUGGAGGUCAAAACCACCGCUAGCACCAAGUUUGGCAUGGAUGCCUUGGGUCAUGCUACCGCCGACGCGGUGACCACCACC .....------------.......((((.((((.........((..((.((((....((((....))))))))))...)).(((((....))))))))).)))). ( -30.50) >consensus AGUGGCGGGAGCCAUCCCGCCGAAGUGGAGGUAACCACCCAGGCUCGAGCCACUUUCGGCAUGGACGCCGAAGGACAUGCUACCGGCGAGGCAGUGACCACCACC ..((((((((....)))))))).(((((.(((.........(((....))).((((((((......)))))))).((((((.(....).))).)))))).))))) (-29.82 = -32.53 + 2.71)


| Location | 18,372,677 – 18,372,788 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 77.75 |
| Mean single sequence MFE | -42.20 |
| Consensus MFE | -28.98 |
| Energy contribution | -30.82 |
| Covariance contribution | 1.84 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.11 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.582741 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18372677 111 + 27905053 UCGGCGUCCAUGCCGAAAGUGGCUCGAGCCUGGGUGGUUACCUCCACCUCGGCGGGAUGGCUCCCGCCACUCACAUCAGCCGCUGAGCCCUGUUGCUCGAGGAGAUUAGUG ((((((....)))))).(((((((.(((((.((((((......)))))).)))(((.((((....)))))))...)))))))))((((......))))............. ( -50.10) >DroSec_CAF1 37163 111 + 1 UCGGCGUCCAUCCCGAAAGUGGCUCGAGCCUGGGUGGUUACCUCCACCUCGGCAGGAUGGCUCCCGCCACUCACAUCAGCCGCUGAGCCCUGUUGCUCCAGAAGAUUGGUG ((((.(.....))))).(((((((.(((((.((((((......)))))).))).(..((((....))))..)...)))))))))((((......))))............. ( -43.90) >DroSim_CAF1 38444 111 + 1 UCGGCGUCCAUCCCGAAAGUGGCUCGAGCCUGAGUGGUUACCUCCACCUCGGCGGGAUGGCUCCCGCCACUCCCAUCAGCCGCUGAGCCCUGUUGCUCCAGAAGAUUGGUG ((((.(.....))))).(((((((.(((((.((((((......))).))))))((((((((....)))).)))).)))))))))((((......))))............. ( -44.40) >DroEre_CAF1 38186 111 + 1 UCGGCGUCCAUGCCAAAAGUGGCACGAGCCUGGGUGGUUACCUCUACUUCGGCGGGAUGGCUCCCGCCACUCACAUCAGCCGCUGAGCCCUGACGCUCCAGGAGAUUUGUG ..((((....))))...((((((....(((..(((((......)))))..)))((((....))))))))))((((....((...((((......))))..)).....)))) ( -42.90) >DroYak_CAF1 38134 96 + 1 UCGGCGUCCAUGCCAAAAGUGGCACGGGCCUGGGUGGUUACCUCCACUUCGGCGG---------------CCACAUCAGCCGCUGAGCCGUGUGGCUCCAGGAGAUUGGUG ..(((..((.(((((....))))).))(((..(((((......)))))..))).)---------------)).(((((.((...(((((....)))))..))....))))) ( -42.40) >DroAna_CAF1 48871 99 + 1 AAGGCAUCCAUGCCAAACUUGGUGCUAGCGGUGGUUUUGACCUCCACUUCAGCA------------UCGUUCAGAUCGUUCGUGGAGUUGUGUCGCUCGAGAAGAUUCGAU ..((((....))))......((((((((.((.(((....))).)).))..))))------------))..((..(((.(((.(.((((......)))).)))))))..)). ( -29.50) >consensus UCGGCGUCCAUGCCAAAAGUGGCUCGAGCCUGGGUGGUUACCUCCACCUCGGCGGGAUGGCUCCCGCCACUCACAUCAGCCGCUGAGCCCUGUUGCUCCAGAAGAUUGGUG ..((((....))))...((((((....(((.((((((......)))))).)))(((.((((....)))))))......))))))((((......))))............. (-28.98 = -30.82 + 1.84)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:34:23 2006