
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,334,796 – 18,334,928 |
| Length | 132 |
| Max. P | 0.999339 |

| Location | 18,334,796 – 18,334,915 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.66 |
| Mean single sequence MFE | -50.90 |
| Consensus MFE | -45.74 |
| Energy contribution | -46.74 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.90 |
| SVM decision value | 3.52 |
| SVM RNA-class probability | 0.999339 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
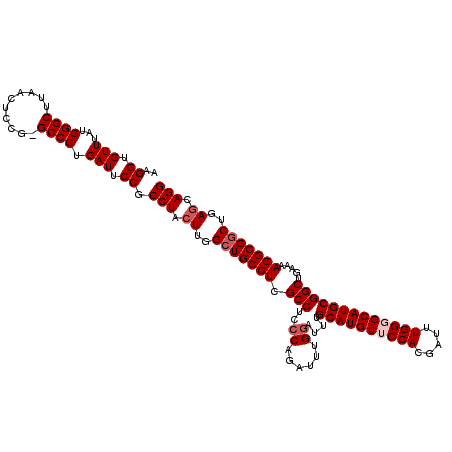
>3R_DroMel_CAF1 18334796 119 + 27905053 AAGGUGUUUAUGGGCUUAACUCCG-GCCCUGAUUCCGCCUACUUGGCUGCCUCGCCUCACAGAUUUGGAUUUGCAUGUGCCACGAUUUGGGGCAUGCGGCUGAGAAGGCGGCUGAGCAGG ..((.(((...(((((.......)-)))).))).)).(((.((..((((((((((((((......)))....((((((.(((.....))).)))))))))...).)))))))..)).))) ( -48.60) >DroSec_CAF1 20947 119 + 1 AAGGUGUUUAUGGGCUUAACUCCG-GCCCUGAUUCCGCCUACUUGGCUGCCUCGCCUCCCAGAUUUGGAUUUGCAUGUUCCACGAUUUGGGGCAUGCGGCUGAAAAGGCGGCUGAGCAGG ..((.(((...(((((.......)-)))).))).)).(((.((..(((((((.(((..((......))....((((((((((.....))))))))))))).....)))))))..)).))) ( -51.80) >DroSim_CAF1 21410 119 + 1 AAGGUGUUUAUGGGCUUAACUCCG-GCCCUGAUUCCGCCUACUUGGCUGCCUCGCCUCCCAGAUUUGGAUUUGCAUGUUCCACGAUUUGGGGCAUGCGGCUGAAAAGGCGGCUGAGCAGG ..((.(((...(((((.......)-)))).))).)).(((.((..(((((((.(((..((......))....((((((((((.....))))))))))))).....)))))))..)).))) ( -51.80) >DroEre_CAF1 21326 120 + 1 AAGGUGUUUAUGGGCUUAACUCCGGGCCCUGAUUCCGCCUACUUGGCUGCCUCGCCUUCCAGAUUUGGAUUUGCAUGUUCCACGAUUUGGGGCAUGCGGCUGAGAAGGCGACUGAACAGG ....((((((.((((((......))))))......(((((.((..(((((...(((..(((((((((((.........)))).))))))))))..)))))..)).)))))..)))))).. ( -50.30) >DroYak_CAF1 22910 120 + 1 AAGGUGUUUAUGGGCUUAACUCCGGGCCCUGAUUCCGCCUACUUGGCUGCCUCGCCUCCCAGAUUUGGAUUUGCAUGUGCCACGAUUUGGGGCAUGCGGCUGACAAGGCGGCUGAGAAGG .(((((.(((.((((((......)))))))))...))))).((..(((((((.(((..((......))....((((((.(((.....))).))))))))).....)))))))..)).... ( -52.00) >consensus AAGGUGUUUAUGGGCUUAACUCCG_GCCCUGAUUCCGCCUACUUGGCUGCCUCGCCUCCCAGAUUUGGAUUUGCAUGUUCCACGAUUUGGGGCAUGCGGCUGAAAAGGCGGCUGAGCAGG ..((.(((...((((..........)))).))).)).(((.((..(((((((.(((..((......))....((((((((((.....))))))))))))).....)))))))..)).))) (-45.74 = -46.74 + 1.00)


| Location | 18,334,796 – 18,334,915 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.66 |
| Mean single sequence MFE | -40.18 |
| Consensus MFE | -38.18 |
| Energy contribution | -38.10 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.970167 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18334796 119 - 27905053 CCUGCUCAGCCGCCUUCUCAGCCGCAUGCCCCAAAUCGUGGCACAUGCAAAUCCAAAUCUGUGAGGCGAGGCAGCCAAGUAGGCGGAAUCAGGGC-CGGAGUUAAGCCCAUAAACACCUU ((((((..((.(((((....((((((((..(((.....)))..)))))...((((....)).)))))))))).))..))))))........((((-.........))))........... ( -38.70) >DroSec_CAF1 20947 119 - 1 CCUGCUCAGCCGCCUUUUCAGCCGCAUGCCCCAAAUCGUGGAACAUGCAAAUCCAAAUCUGGGAGGCGAGGCAGCCAAGUAGGCGGAAUCAGGGC-CGGAGUUAAGCCCAUAAACACCUU ((((((..((.(((((....((((((((..(((.....)))..)))))...((((....)))).)))))))).))..))))))........((((-.........))))........... ( -41.70) >DroSim_CAF1 21410 119 - 1 CCUGCUCAGCCGCCUUUUCAGCCGCAUGCCCCAAAUCGUGGAACAUGCAAAUCCAAAUCUGGGAGGCGAGGCAGCCAAGUAGGCGGAAUCAGGGC-CGGAGUUAAGCCCAUAAACACCUU ((((((..((.(((((....((((((((..(((.....)))..)))))...((((....)))).)))))))).))..))))))........((((-.........))))........... ( -41.70) >DroEre_CAF1 21326 120 - 1 CCUGUUCAGUCGCCUUCUCAGCCGCAUGCCCCAAAUCGUGGAACAUGCAAAUCCAAAUCUGGAAGGCGAGGCAGCCAAGUAGGCGGAAUCAGGGCCCGGAGUUAAGCCCAUAAACACCUU (((((((....(((((....((((((((..(((.....)))..)))))...((((....)))).)))))))).(((.....))).))).))))....((.(((.........))).)).. ( -38.90) >DroYak_CAF1 22910 120 - 1 CCUUCUCAGCCGCCUUGUCAGCCGCAUGCCCCAAAUCGUGGCACAUGCAAAUCCAAAUCUGGGAGGCGAGGCAGCCAAGUAGGCGGAAUCAGGGCCCGGAGUUAAGCCCAUAAACACCUU .........((((((((((....(((((..(((.....)))..)))))...((((....)))).)))))))).(((.....))))).....((((..........))))........... ( -39.90) >consensus CCUGCUCAGCCGCCUUCUCAGCCGCAUGCCCCAAAUCGUGGAACAUGCAAAUCCAAAUCUGGGAGGCGAGGCAGCCAAGUAGGCGGAAUCAGGGC_CGGAGUUAAGCCCAUAAACACCUU ((((((..((.(((((....((((((((..(((.....)))..)))))...((((....)))).)))))))).))..))))))........((((..........))))........... (-38.18 = -38.10 + -0.08)


| Location | 18,334,835 – 18,334,928 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 93.89 |
| Mean single sequence MFE | -36.92 |
| Consensus MFE | -32.56 |
| Energy contribution | -33.56 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.92 |
| SVM RNA-class probability | 0.997714 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18334835 93 + 27905053 ACUUGGCUGCCUCGCCUCACAGAUUUGGAUUUGCAUGUGCCACGAUUUGGGGCAUGCGGCUGAGAAGGCGGCUGAGCAGGCG-----GAUAGAAGACA .((..((((((((((((((......)))....((((((.(((.....))).)))))))))...).)))))))..))......-----........... ( -33.90) >DroSec_CAF1 20986 93 + 1 ACUUGGCUGCCUCGCCUCCCAGAUUUGGAUUUGCAUGUUCCACGAUUUGGGGCAUGCGGCUGAAAAGGCGGCUGAGCAGGCG-----GAUAGAAGACA .((..(((((((.(((..((......))....((((((((((.....))))))))))))).....)))))))..))......-----........... ( -37.10) >DroSim_CAF1 21449 93 + 1 ACUUGGCUGCCUCGCCUCCCAGAUUUGGAUUUGCAUGUUCCACGAUUUGGGGCAUGCGGCUGAAAAGGCGGCUGAGCAGGCG-----GAUAGAAGACA .((..(((((((.(((..((......))....((((((((((.....))))))))))))).....)))))))..))......-----........... ( -37.10) >DroEre_CAF1 21366 93 + 1 ACUUGGCUGCCUCGCCUUCCAGAUUUGGAUUUGCAUGUUCCACGAUUUGGGGCAUGCGGCUGAGAAGGCGACUGAACAGGCA-----GAUAGAAGACA .((...((((((((((((((((........((((((((((((.....))))))))))))))).)))))))).......))))-----)..))...... ( -38.81) >DroYak_CAF1 22950 98 + 1 ACUUGGCUGCCUCGCCUCCCAGAUUUGGAUUUGCAUGUGCCACGAUUUGGGGCAUGCGGCUGACAAGGCGGCUGAGAAGGCGACUGGGAUAGAAGACA .((..(((((((.(((..((......))....((((((.(((.....))).))))))))).....)))))))..)).((....))............. ( -37.70) >consensus ACUUGGCUGCCUCGCCUCCCAGAUUUGGAUUUGCAUGUUCCACGAUUUGGGGCAUGCGGCUGAAAAGGCGGCUGAGCAGGCG_____GAUAGAAGACA .((..(((((((.(((..((......))....((((((((((.....))))))))))))).....)))))))..))...................... (-32.56 = -33.56 + 1.00)



| Location | 18,334,835 – 18,334,928 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 93.89 |
| Mean single sequence MFE | -27.94 |
| Consensus MFE | -25.44 |
| Energy contribution | -25.36 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.717487 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 18334835 93 - 27905053 UGUCUUCUAUC-----CGCCUGCUCAGCCGCCUUCUCAGCCGCAUGCCCCAAAUCGUGGCACAUGCAAAUCCAAAUCUGUGAGGCGAGGCAGCCAAGU ...........-----.....(((..((.(((((....((((((((..(((.....)))..)))))...((((....)).)))))))))).))..))) ( -25.10) >DroSec_CAF1 20986 93 - 1 UGUCUUCUAUC-----CGCCUGCUCAGCCGCCUUUUCAGCCGCAUGCCCCAAAUCGUGGAACAUGCAAAUCCAAAUCUGGGAGGCGAGGCAGCCAAGU ...........-----.....(((..((.(((((....((((((((..(((.....)))..)))))...((((....)))).)))))))).))..))) ( -28.10) >DroSim_CAF1 21449 93 - 1 UGUCUUCUAUC-----CGCCUGCUCAGCCGCCUUUUCAGCCGCAUGCCCCAAAUCGUGGAACAUGCAAAUCCAAAUCUGGGAGGCGAGGCAGCCAAGU ...........-----.....(((..((.(((((....((((((((..(((.....)))..)))))...((((....)))).)))))))).))..))) ( -28.10) >DroEre_CAF1 21366 93 - 1 UGUCUUCUAUC-----UGCCUGUUCAGUCGCCUUCUCAGCCGCAUGCCCCAAAUCGUGGAACAUGCAAAUCCAAAUCUGGAAGGCGAGGCAGCCAAGU ..........(-----((((.......((((((((.(((..(((((..(((.....)))..)))))..........))))))))))))))))...... ( -29.11) >DroYak_CAF1 22950 98 - 1 UGUCUUCUAUCCCAGUCGCCUUCUCAGCCGCCUUGUCAGCCGCAUGCCCCAAAUCGUGGCACAUGCAAAUCCAAAUCUGGGAGGCGAGGCAGCCAAGU ......................((..((.((((((((....(((((..(((.....)))..)))))...((((....)))).)))))))).))..)). ( -29.30) >consensus UGUCUUCUAUC_____CGCCUGCUCAGCCGCCUUCUCAGCCGCAUGCCCCAAAUCGUGGAACAUGCAAAUCCAAAUCUGGGAGGCGAGGCAGCCAAGU .....................(((..((.(((((....((((((((..(((.....)))..)))))...((((....)))).)))))))).))..))) (-25.44 = -25.36 + -0.08)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:34:19 2006