| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 18,328,943 – 18,329,041 |
| Length | 98 |
| Max. P | 0.627247 |

| Location | 18,328,943 – 18,329,041 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.16 |
| Mean single sequence MFE | -21.95 |
| Consensus MFE | -16.51 |
| Energy contribution | -16.77 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.12 |
| Mean z-score | -0.93 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.627247 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
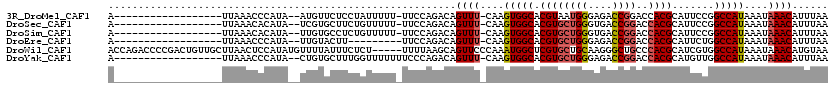
>3R_DroMel_CAF1 18328943 98 - 27905053 A------------------UUAAACCCAUA--AUGUUCUCCUAUUUUU-UUCCAGACAGUUU-CAAGUGGCACGUAAUGGGAGACCGGACCACGCAUUCCGGCCAUAAAUAAACAUUUAA .------------------..........(--(((((((((((((...-..(((........-....))).....)))))))))(((((........)))))..........)))))... ( -20.20) >DroSec_CAF1 14474 98 - 1 A------------------UUAAACACAUA--UCGUGCUUCUGUUUUU-UUCCAGACAGUUU-CAAGUGGCACGUGCUGGGUGACCGGACCACGCAUUCCGGCCAUAAAUAAACAUUUAA .------------------.......((..--.((((((.((((((..-....))))))...-.....))))))...)).(((.(((((........))))).))).............. ( -22.10) >DroSim_CAF1 14740 98 - 1 A------------------UUAAACACAUA--UUGUGCCUCUGUUUUU-UUCCAGACAGUUU-CAAGUGGCACGUGCUGGGUGACCGGACCACGCAUUCCGGCCAUAAAUAAACAUUUAA .------------------...........--..(((((.((((((..-....))))))...-.....)))))((.....(((.(((((........))))).)))......))...... ( -22.70) >DroEre_CAF1 14872 90 - 1 A------------------UUAAACCCAUA--UUGUACUU---------UUCCAGACAGUUU-CAAGUGGCACGUGCUGGGAGACCGGACCACGCAUUCUGGCCAUAAAUAAACAUUUAA .------------------..........(--((((.((.---------....)))))))..-...(((((.((((((((....))))..)))).......))))).............. ( -18.30) >DroWil_CAF1 15549 115 - 1 ACCAGACCCCGACUGUUGCUUAACUCCAUAUGUUUUAUUUCUCU-----UUUUAAGCAGUUCCCAAAUGGCUCGUGCUGCAAGGGCUGCCCACGCAUCGUGGCCAUAAAUAAACAUGUAA ..(((.......)))............((((((((.((((....-----......((((..(((...((((....))))...)))))))(((((...)))))....)))))))))))).. ( -22.30) >DroYak_CAF1 15340 99 - 1 A------------------UUAAACCCAUA--CUGUGCUUUGGUUUUUUUCCCAGACAGUUU-CAAGUGGCACGUGCUGGGAGACCGGACCACGCAUGUUGGCCAUAAAUAAACAUUUAA .------------------......(((.(--(.((((..((((((.((((((((...((.(-(....)).))...))))))))..)))))).))))))))).................. ( -26.10) >consensus A__________________UUAAACCCAUA__UUGUGCUUCUGUUUUU_UUCCAGACAGUUU_CAAGUGGCACGUGCUGGGAGACCGGACCACGCAUUCCGGCCAUAAAUAAACAUUUAA ..........................................................((((....(((((.((((((((....))))..)))).......)))))....))))...... (-16.51 = -16.77 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:34:06 2006